Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Prespliceosome structure provides insights into spliceosome assembly and regulation
Nature ( IF 50.5 ) Pub Date : 2018-07-01 , DOI: 10.1038/s41586-018-0323-8 Clemens Plaschka 1, 2 , Pei-Chun Lin 1 , Clément Charenton 1 , Kiyoshi Nagai 1
Nature ( IF 50.5 ) Pub Date : 2018-07-01 , DOI: 10.1038/s41586-018-0323-8 Clemens Plaschka 1, 2 , Pei-Chun Lin 1 , Clément Charenton 1 , Kiyoshi Nagai 1
Affiliation
|
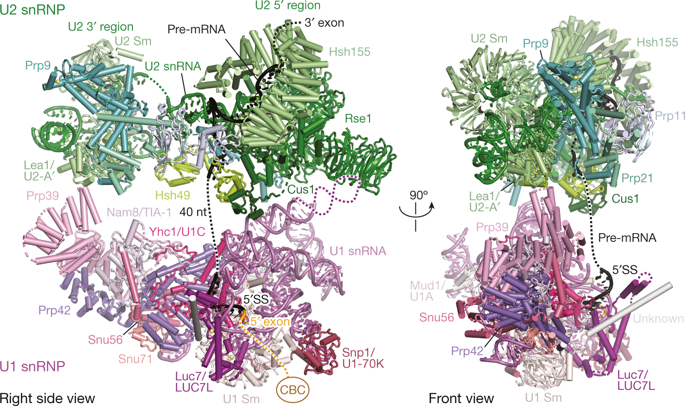
The spliceosome catalyses the excision of introns from pre-mRNA in two steps, branching and exon ligation, and is assembled from five small nuclear ribonucleoprotein particles (snRNPs; U1, U2, U4, U5, U6) and numerous non-snRNP factors1. For branching, the intron 5′ splice site and the branch point sequence are selected and brought by the U1 and U2 snRNPs into the prespliceosome1, which is a focal point for regulation by alternative splicing factors2. The U4/U6.U5 tri-snRNP subsequently joins the prespliceosome to form the complete pre-catalytic spliceosome. Recent studies have revealed the structural basis of the branching and exon-ligation reactions3, however, the structural basis of the early events in spliceosome assembly remains poorly understood4. Here we report the cryo-electron microscopy structure of the yeast Saccharomyces cerevisiae prespliceosome at near-atomic resolution. The structure reveals an induced stabilization of the 5′ splice site in the U1 snRNP, and provides structural insights into the functions of the human alternative splicing factors LUC7-like (yeast Luc7) and TIA-1 (yeast Nam8), both of which have been linked to human disease5,6. In the prespliceosome, the U1 snRNP associates with the U2 snRNP through a stable contact with the U2 3′ domain and a transient yeast-specific contact with the U2 SF3b-containing 5′ region, leaving its tri-snRNP-binding interface fully exposed. The results suggest mechanisms for 5′ splice site transfer to the U6 ACAGAGA region within the assembled spliceosome and for its subsequent conversion to the activation-competent B-complex spliceosome7,8. Taken together, the data provide a working model to investigate the early steps of spliceosome assembly.The cryo-electron microscopy structure of the Saccharomyces cerevisiae prespliceosome provides insights into splice-site selection and early spliceosome assembly events.
中文翻译:

Prespliceosome 结构提供了对剪接体组装和调控的见解
剪接体分两步催化从前 mRNA 中切除内含子,分支和外显子连接,并由五个小的核核糖核蛋白颗粒(snRNP;U1、U2、U4、U5、U6)和许多非 snRNP 因子组装而成。对于分支,内含子 5' 剪接位点和分支点序列被 U1 和 U2 snRNP 选择并带入前剪接体 1,这是可变剪接因子调节的焦点 2。U4/U6.U5 tri-snRNP 随后加入前剪接体以形成完整的预催化剪接体。最近的研究揭示了分支和外显子连接反应的结构基础,然而,剪接体组装中早期事件的结构基础仍然知之甚少。在这里,我们报告了酵母酿酒酵母前剪接体在近原子分辨率下的低温电子显微镜结构。该结构揭示了 U1 snRNP 中 5' 剪接位点的诱导稳定化,并为人类选择性剪接因子 LUC7 样(酵母 Luc7)和 TIA-1(酵母 Nam8)的功能提供了结构性见解,两者都具有与人类疾病有关5,6。在前剪接体中,U1 snRNP 通过与 U2 3' 结构域的稳定接触和与含有 U2 SF3b 的 5' 区域的瞬时酵母特异性接触与 U2 snRNP 结合,使其三-snRNP 结合界面完全暴露。结果表明 5' 剪接位点转移到组装剪接体内的 U6 ACAGAGA 区域以及随后转化为具有活化能力的 B 复合体剪接体的机制7, 8。总之,这些数据为研究剪接体组装的早期步骤提供了一个工作模型。酿酒酵母前剪接体的低温电子显微镜结构提供了对剪接位点选择和早期剪接体组装事件的见解。
更新日期:2018-07-01
中文翻译:

Prespliceosome 结构提供了对剪接体组装和调控的见解
剪接体分两步催化从前 mRNA 中切除内含子,分支和外显子连接,并由五个小的核核糖核蛋白颗粒(snRNP;U1、U2、U4、U5、U6)和许多非 snRNP 因子组装而成。对于分支,内含子 5' 剪接位点和分支点序列被 U1 和 U2 snRNP 选择并带入前剪接体 1,这是可变剪接因子调节的焦点 2。U4/U6.U5 tri-snRNP 随后加入前剪接体以形成完整的预催化剪接体。最近的研究揭示了分支和外显子连接反应的结构基础,然而,剪接体组装中早期事件的结构基础仍然知之甚少。在这里,我们报告了酵母酿酒酵母前剪接体在近原子分辨率下的低温电子显微镜结构。该结构揭示了 U1 snRNP 中 5' 剪接位点的诱导稳定化,并为人类选择性剪接因子 LUC7 样(酵母 Luc7)和 TIA-1(酵母 Nam8)的功能提供了结构性见解,两者都具有与人类疾病有关5,6。在前剪接体中,U1 snRNP 通过与 U2 3' 结构域的稳定接触和与含有 U2 SF3b 的 5' 区域的瞬时酵母特异性接触与 U2 snRNP 结合,使其三-snRNP 结合界面完全暴露。结果表明 5' 剪接位点转移到组装剪接体内的 U6 ACAGAGA 区域以及随后转化为具有活化能力的 B 复合体剪接体的机制7, 8。总之,这些数据为研究剪接体组装的早期步骤提供了一个工作模型。酿酒酵母前剪接体的低温电子显微镜结构提供了对剪接位点选择和早期剪接体组装事件的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号