Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks
Nature ( IF 50.5 ) Pub Date : 2018-07-01 , DOI: 10.1038/s41586-018-0289-6 Kevin M. Cherry , Lulu Qian
Nature ( IF 50.5 ) Pub Date : 2018-07-01 , DOI: 10.1038/s41586-018-0289-6 Kevin M. Cherry , Lulu Qian
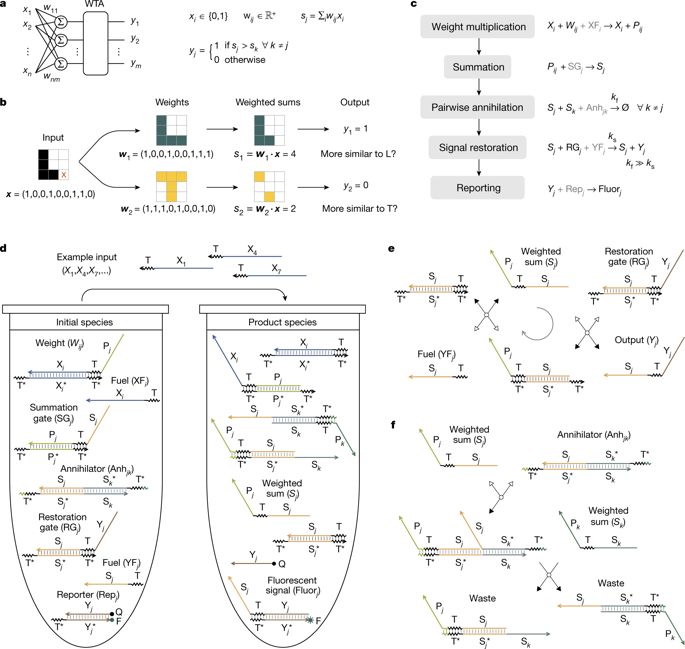
|
From bacteria following simple chemical gradients1 to the brain distinguishing complex odour information2, the ability to recognize molecular patterns is essential for biological organisms. This type of information-processing function has been implemented using DNA-based neural networks3, but has been limited to the recognition of a set of no more than four patterns, each composed of four distinct DNA molecules. Winner-take-all computation4 has been suggested5,6 as a potential strategy for enhancing the capability of DNA-based neural networks. Compared to the linear-threshold circuits7 and Hopfield networks8 used previously3, winner-take-all circuits are computationally more powerful4, allow simpler molecular implementation and are not constrained by the number of patterns and their complexity, so both a large number of simple patterns and a small number of complex patterns can be recognized. Here we report a systematic implementation of winner-take-all neural networks based on DNA-strand-displacement9,10 reactions. We use a previously developed seesaw DNA gate motif3,11,12, extended to include a simple and robust component that facilitates the cooperative hybridization13 that is involved in the process of selecting a ‘winner’. We show that with this extended seesaw motif DNA-based neural networks can classify patterns into up to nine categories. Each of these patterns consists of 20 distinct DNA molecules chosen from the set of 100 that represents the 100 bits in 10 × 10 patterns, with the 20 DNA molecules selected tracing one of the handwritten digits ‘1’ to ‘9’. The network successfully classified test patterns with up to 30 of the 100 bits flipped relative to the digit patterns ‘remembered’ during training, suggesting that molecular circuits can robustly accomplish the sophisticated task of classifying highly complex and noisy information on the basis of similarity to a memory.DNA-strand-displacement reactions are used to implement a neural network that can distinguish complex and noisy molecular patterns from a set of nine possibilities—an improvement on previous demonstrations that distinguished only four simple patterns.
中文翻译:

使用基于 DNA 的赢家通吃神经网络扩大分子模式识别
从遵循简单化学梯度的细菌 1 到大脑区分复杂气味信息 2,识别分子模式的能力对于生物有机体至关重要。这种类型的信息处理功能已使用基于 DNA 的神经网络实现,但仅限于识别一组不超过四种模式,每种模式由四种不同的 DNA 分子组成。赢家通吃计算 4 已被建议 5,6 作为增强基于 DNA 的神经网络能力的潜在策略。与之前使用的线性阈值电路 7 和 Hopfield 网络 8 相比,赢家通吃电路在计算上更强大 4,允许更简单的分子实现,并且不受模式数量及其复杂性的限制,所以大量的简单图案和少量的复杂图案都可以识别。在这里,我们报告了基于 DNA 链置换 9,10 反应的赢家通吃神经网络的系统实现。我们使用先前开发的跷跷板 DNA 门基序 3、11、12,扩展为包括一个简单而强大的组件,可促进选择“赢家”过程中涉及的合作杂交 13。我们表明,通过这种扩展的跷跷板基序,基于 DNA 的神经网络可以将模式分为多达九类。这些模式中的每一个都由 20 个不同的 DNA 分子组成,这些分子从 100 个一组中选出,代表 10 × 10 模式中的 100 位,选择的 20 个 DNA 分子追踪手写数字“1”到“9”之一。
更新日期:2018-07-01
中文翻译:

使用基于 DNA 的赢家通吃神经网络扩大分子模式识别
从遵循简单化学梯度的细菌 1 到大脑区分复杂气味信息 2,识别分子模式的能力对于生物有机体至关重要。这种类型的信息处理功能已使用基于 DNA 的神经网络实现,但仅限于识别一组不超过四种模式,每种模式由四种不同的 DNA 分子组成。赢家通吃计算 4 已被建议 5,6 作为增强基于 DNA 的神经网络能力的潜在策略。与之前使用的线性阈值电路 7 和 Hopfield 网络 8 相比,赢家通吃电路在计算上更强大 4,允许更简单的分子实现,并且不受模式数量及其复杂性的限制,所以大量的简单图案和少量的复杂图案都可以识别。在这里,我们报告了基于 DNA 链置换 9,10 反应的赢家通吃神经网络的系统实现。我们使用先前开发的跷跷板 DNA 门基序 3、11、12,扩展为包括一个简单而强大的组件,可促进选择“赢家”过程中涉及的合作杂交 13。我们表明,通过这种扩展的跷跷板基序,基于 DNA 的神经网络可以将模式分为多达九类。这些模式中的每一个都由 20 个不同的 DNA 分子组成,这些分子从 100 个一组中选出,代表 10 × 10 模式中的 100 位,选择的 20 个 DNA 分子追踪手写数字“1”到“9”之一。











































 京公网安备 11010802027423号
京公网安备 11010802027423号