当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular Approaches To Address the Challenges of RNA Analysis in Complex Matrices
Analytical Chemistry ( IF 6.7 ) Pub Date : 2018-07-03 00:00:00 , DOI: 10.1021/acs.analchem.8b01621 Shan S. Lansing 1 , Susmitha Matlapudi 1 , Sean M. Burrows 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2018-07-03 00:00:00 , DOI: 10.1021/acs.analchem.8b01621 Shan S. Lansing 1 , Susmitha Matlapudi 1 , Sean M. Burrows 1
Affiliation
|
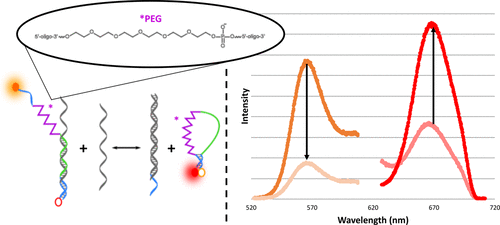
We present on a design change and addition of an internal polyethylene glycol (PEG) spacer to an existing biosensor. There were two reasons for changing the sensor design. The first was to increase the stability of the biosensor to avoid binding off-analytes with single nucleotide polymorphisms. The second was to prevent sensor degradation by nucleases. The biosensor, designed for detection of short noncoding RNA strands, is composed of Reporter and Probe nucleic acid strands that form a partially complementary duplex. The internal PEG was added to the Reporter, and subsequently diminished false negatives that resulted from off-oligonucleotide binding. Furthermore, the PEG eliminated degradation of the sensor by DNase1 endonuclease. Currently, in situ and crude cell lysate RNA analysis is hindered by nonspecific interactions and degradation by endogenous nucleases. Together, the design changes presented here mitigate these matrix effects and allow for robust RNA analysis in complex media.
中文翻译:

解决复杂基质中RNA分析挑战的分子方法
我们提出了设计更改,并向现有的生物传感器添加了内部聚乙二醇(PEG)间隔物。更改传感器设计有两个原因。第一个是增加生物传感器的稳定性,以避免与单核苷酸多态性结合脱分析物。第二是防止核酸酶降解传感器。该生物传感器设计用于检测短的非编码RNA链,由组成部分互补双链体的Reporter和Probe核酸链组成。内部PEG被添加到Reporter中,随后减少了由寡核苷酸结合产生的假阴性。此外,PEG消除了DNase1核酸内切酶对传感器的降解。目前,非特异性相互作用和内源核酸酶降解阻碍了原位和粗细胞裂解物RNA分析。总之,此处介绍的设计更改减轻了这些基质效应,并允许在复杂介质中进行可靠的RNA分析。
更新日期:2018-07-03
中文翻译:

解决复杂基质中RNA分析挑战的分子方法
我们提出了设计更改,并向现有的生物传感器添加了内部聚乙二醇(PEG)间隔物。更改传感器设计有两个原因。第一个是增加生物传感器的稳定性,以避免与单核苷酸多态性结合脱分析物。第二是防止核酸酶降解传感器。该生物传感器设计用于检测短的非编码RNA链,由组成部分互补双链体的Reporter和Probe核酸链组成。内部PEG被添加到Reporter中,随后减少了由寡核苷酸结合产生的假阴性。此外,PEG消除了DNase1核酸内切酶对传感器的降解。目前,非特异性相互作用和内源核酸酶降解阻碍了原位和粗细胞裂解物RNA分析。总之,此处介绍的设计更改减轻了这些基质效应,并允许在复杂介质中进行可靠的RNA分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号