Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High speed of fork progression induces DNA replication stress and genomic instability
Nature ( IF 50.5 ) Pub Date : 2018-06-27 , DOI: 10.1038/s41586-018-0261-5 Apolinar Maya-Mendoza , Pavel Moudry , Joanna Maria Merchut-Maya , MyungHee Lee , Robert Strauss , Jiri Bartek
Nature ( IF 50.5 ) Pub Date : 2018-06-27 , DOI: 10.1038/s41586-018-0261-5 Apolinar Maya-Mendoza , Pavel Moudry , Joanna Maria Merchut-Maya , MyungHee Lee , Robert Strauss , Jiri Bartek
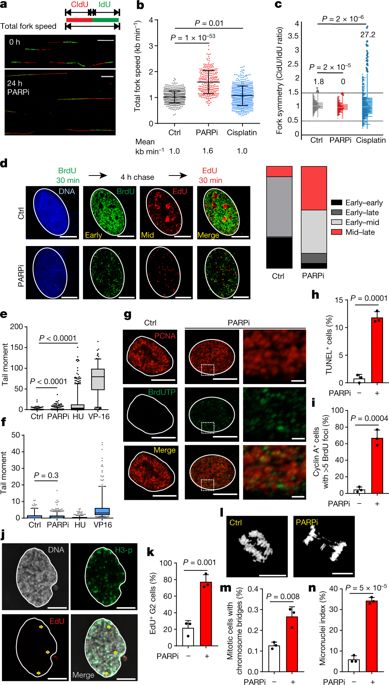
|
Accurate replication of DNA requires stringent regulation to ensure genome integrity. In human cells, thousands of origins of replication are coordinately activated during S phase, and the velocity of replication forks is adjusted to fully replicate DNA in pace with the cell cycle1. Replication stress induces fork stalling and fuels genome instability2. The mechanistic basis of replication stress remains poorly understood despite its emerging role in promoting cancer2. Here we show that inhibition of poly(ADP-ribose) polymerase (PARP) increases the speed of fork elongation and does not cause fork stalling, which is in contrast to the accepted model in which inhibitors of PARP induce fork stalling and collapse3. Aberrant acceleration of fork progression by 40% above the normal velocity leads to DNA damage. Depletion of the treslin or MTBP proteins, which are involved in origin firing, also increases fork speed above the tolerated threshold, and induces the DNA damage response pathway. Mechanistically, we show that poly(ADP-ribosyl)ation (PARylation) and the PCNA interactor p21Cip1 (p21) are crucial modulators of fork progression. PARylation and p21 act as suppressors of fork speed in a coordinated regulatory network that is orchestrated by the PARP1 and p53 proteins. Moreover, at the fork level, PARylation acts as a sensor of replication stress. During PARP inhibition, DNA lesions that induce fork arrest and are normally resolved or repaired remain unrecognized by the replication machinery. Conceptually, our results show that accelerated replication fork progression represents a general mechanism that triggers replication stress and the DNA damage response. Our findings contribute to a better understanding of the mechanism of fork speed control, with implications for genomic (in)stability and rational cancer treatment.Inhibition of PARP is shown to accelerate the speed of replication fork elongation, which prevents fork stalling and induces DNA damage, with implications for genomic instability and cancer treatment.
中文翻译:

高速叉进展诱导 DNA 复制压力和基因组不稳定性
DNA 的准确复制需要严格的监管以确保基因组的完整性。在人体细胞中,在S期协调激活数千个复制起点,并调整复制叉的速度,以与细胞周期同步完全复制DNA1。复制压力会导致叉停顿并加剧基因组不稳定2。尽管复制应激在促进癌症方面发挥了新的作用,但对复制应激的机制基础仍然知之甚少。在这里,我们表明抑制聚(ADP-核糖)聚合酶 (PARP) 会增加叉子伸长的速度,并且不会导致叉子停顿,这与 PARP 抑制剂诱导叉子停顿和坍塌的公认模型形成对比。前叉进展的异常加速比正常速度高 40% 会导致 DNA 损伤。treslin 或 MTBP 蛋白的消耗,参与原点激发,也将叉速度提高到耐受阈值以上,并诱导 DNA 损伤反应途径。从机制上讲,我们表明聚(ADP-核糖基)化 (PARylation) 和 PCNA 相互作用因子 p21Cip1 (p21) 是叉进展的关键调节剂。PARylation 和 p21 在由 PARP1 和 p53 蛋白协调的协调调节网络中充当叉速度的抑制剂。此外,在 fork 级别,PARylation 充当复制压力的传感器。在 PARP 抑制期间,诱导叉停止并正常解决或修复的 DNA 损伤仍未被复制机制识别。从概念上讲,我们的结果表明,加速复制叉进展代表了触发复制压力和 DNA 损伤反应的一般机制。
更新日期:2018-06-27
中文翻译:

高速叉进展诱导 DNA 复制压力和基因组不稳定性
DNA 的准确复制需要严格的监管以确保基因组的完整性。在人体细胞中,在S期协调激活数千个复制起点,并调整复制叉的速度,以与细胞周期同步完全复制DNA1。复制压力会导致叉停顿并加剧基因组不稳定2。尽管复制应激在促进癌症方面发挥了新的作用,但对复制应激的机制基础仍然知之甚少。在这里,我们表明抑制聚(ADP-核糖)聚合酶 (PARP) 会增加叉子伸长的速度,并且不会导致叉子停顿,这与 PARP 抑制剂诱导叉子停顿和坍塌的公认模型形成对比。前叉进展的异常加速比正常速度高 40% 会导致 DNA 损伤。treslin 或 MTBP 蛋白的消耗,参与原点激发,也将叉速度提高到耐受阈值以上,并诱导 DNA 损伤反应途径。从机制上讲,我们表明聚(ADP-核糖基)化 (PARylation) 和 PCNA 相互作用因子 p21Cip1 (p21) 是叉进展的关键调节剂。PARylation 和 p21 在由 PARP1 和 p53 蛋白协调的协调调节网络中充当叉速度的抑制剂。此外,在 fork 级别,PARylation 充当复制压力的传感器。在 PARP 抑制期间,诱导叉停止并正常解决或修复的 DNA 损伤仍未被复制机制识别。从概念上讲,我们的结果表明,加速复制叉进展代表了触发复制压力和 DNA 损伤反应的一般机制。











































 京公网安备 11010802027423号
京公网安备 11010802027423号