Molecular Catalysis ( IF 3.9 ) Pub Date : 2018-06-26 , DOI: 10.1016/j.mcat.2018.06.013 Qiang Cheng , Qi Chen , Jian-He Xu , Hui-Lei Yu
|
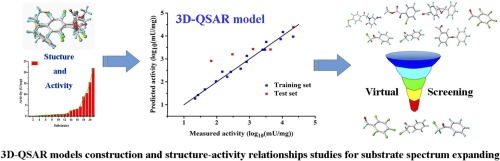
Aldo-keto reductases (AKRs) are widely used for reducing prochiral aldehyde or ketone compounds into corresponding chiral alcohols. In our previous work, an aldo-keto reductase named YtbE was isolated and identified from Bacillus sp. ECU0013, which can catalyze an NADPH-dependent carbonyl reduction reaction with high stereoselectivity. In order to further expand the application range of the enzyme, three-Dimensional Quantitative Structure Activity Relationship (3D-QSAR) models were constructed to predict the specific activity on the aldehyde or ketone compounds. For Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) models, both the cross-validation coefficient (Q2) (0.623, 0.601) are satisfied, suggesting that the models are robust and effective for prediction. The detailed structure-function relationships between the protein YtbE and different substrates were then examined by the contour maps analysis. We found that electrostatic and hydrophobic force molecular fields play the most important roles in determining the catalytic activity, with contribution factors of 0.378 and 0.384, respectively. Furthermore, virtual screening by using the ZINC15 database was performed to expand investigate the substrate spectrum of YtbE. Among the 77 potential substrates predicted, five were experimentally verified, indicating the better accuracy of CoMSIA-molecule model. The 3D-QSAR assisted method was proved to be helpful for rational investigating and expanding of the YtbE substrate spectrum.
中文翻译:

3D-QSAR辅助活性预测策略,用于扩展醛酮还原酶的底物谱
醛基酮还原酶(AKR)被广泛用于将前手性醛或酮化合物还原为相应的手性醇。在我们之前的工作中,从芽孢杆菌属中分离并鉴定了一种名为YtbE的醛酮还原酶。ECU0013,它可以以高的立体选择性催化NADPH依赖的羰基还原反应。为了进一步扩大酶的应用范围,构建了三维定量结构活性关系(3D-QSAR)模型来预测对醛或酮化合物的比活性。对于比较分子场分析(CoMFA)和比较分子相似性指标分析(CoMSIA)模型,交叉验证系数(Q 2)(0.623,0.601)是令人满意的,这表明该模型对于预测而言是鲁棒且有效的。然后通过等高线图分析检查了蛋白质YtbE与不同底物之间的详细结构-功能关系。我们发现静电和疏水力分子场在确定催化活性中起着最重要的作用,贡献因子分别为0.378和0.384。此外,使用ZINC15数据库进行了虚拟筛选,以扩展研究YtbE的底物光谱。在预测的77种潜在底物中,有5种经过实验验证,表明CoMSIA分子模型的准确性更高。事实证明,3D-QSAR辅助方法有助于合理研究和扩展YtbE底物谱。











































 京公网安备 11010802027423号
京公网安备 11010802027423号