Biochimie ( IF 3.3 ) Pub Date : 2018-06-26 , DOI: 10.1016/j.biochi.2018.06.012 Elena S. Dyakonova , Vladimir V. Koval , Alexander A. Lomzov , Alexander A. Ishchenko , Olga S. Fedorova
|
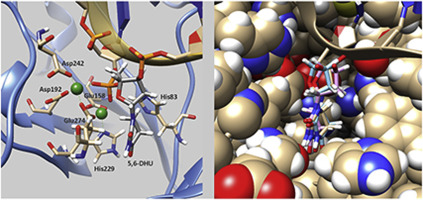
Apurinic/apyrimidinic endonuclease Apn1 of Saccharomyces cerevisiae is known as a key player of the base excision DNA repair (BER) pathway in yeast. BER is initiated by DNA glycosylases, whereas Apn1 can start DNA repair individually in the nucleotide incision repair (NIR) pathway. The aim of this research was to elucidate kinetic and structural dynamic aspects of Apn1 involvement in the NIR process. One of the key characteristics of AP endonuclease's interactions is known to be divalent metal ions playing a part of a cofactor. Well-studied human APE1 employs Mg2+ ions, with metal ion concentration's affecting enzymatic activity exerted by APE1. In our study, we aimed to test the effect of the Mg2+ ion on Apn1's NIR catalysis by examining structural dynamics of DNA during the interaction in real time using the stopped-flow technique. To test NIR activity of Apn1, deoxyribooligonucleotide duplexes containing a 5,6-dihydro-2′-deoxyuridine (DHU) residue were employed as substrates. A 2-aminopurine (2-aPu) residue was a reporter group fluorescence intensity of which was detected during Apn1–DNA interactions. NIR activity of both WT and H83A Apn1 was found to be arrested during the interaction with a DNA duplex containing the 2-aPu residue upstream of DHU. We conducted molecular dynamics simulations to elucidate the structural features of complexes of the enzyme with DHU-containing DNAs. The NIR recruiting S. cerevisiae Apn1 proceeds via multistep rearrangements of the complex of Apn1 with a DHU-containing DNA substrate and results in the incised product of the reaction. For wild-type Apn1, the catalytic rate constants do not depend on the Mg2+ concentration, i.e., they are equal in NIR and BER buffers, with equilibrium association constant Ka being 10-fold higher in NIR buffer.
Our data reveal more delicate regulation of Apn1's NIR activity due to the more complicated kinetic mechanism, as compared to BER.
中文翻译:

来自酿酒酵母的apurinic / apyrimidinic核酸内切酶Apn1被募集到核苷酸切口修复途径:动力学和结构特征
酿酒酵母的apurinic / apyrimidinic核酸内切酶Apn1是酵母中碱基切除DNA修复(BER)途径的关键角色。BER是由DNA糖基化酶引发的,而Apn1可以通过核苷酸切口修复(NIR)途径单独启动DNA修复。这项研究的目的是阐明Apn1参与近红外过程的动力学和结构动力学方面。已知AP核酸内切酶相互作用的关键特征之一是充当辅因子一部分的二价金属离子。经过深入研究的人APE1使用Mg 2+离子,金属离子浓度会影响APE1发挥的酶促活性。在我们的研究中,我们旨在测试Mg 2+的作用。通过使用停流技术实时检查相互作用过程中DNA的结构动力学来分析Apn1的NIR催化离子。为了测试Apn1的NIR活性,将含有5,6-二氢-2'-脱氧尿苷(DHU)残基的脱氧核糖寡核苷酸双链体用作底物。一个2-氨基嘌呤(2-aPu)残基是一个报告基团,其荧光强度在Apn1-DNA相互作用期间被检测到。发现WT和H83A Apn1的NIR活性在与包含DHU上游2-aPu残基的DNA双链体相互作用期间被阻止。我们进行了分子动力学模拟,以阐明该酶与含DHU的DNA的复合物的结构特征。NIR招募酿酒酵母Apn1通过Apn1与含有DHU的DNA底物的复合物的多步重排进行,并产生了反应的产物。对于野生型Apn1,催化速率常数不取决于Mg 2+浓度,即,它们在NIR和BER缓冲液中相等,而平衡缔合常数K a在NIR缓冲液中高10倍。
与BER相比,由于更复杂的动力学机制,我们的数据揭示了对Apn1 NIR活性的更精细调节。











































 京公网安备 11010802027423号
京公网安备 11010802027423号