当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploring the Nonconserved Sequence Space of Synthetic Expression Modules in Bacillus subtilis
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-06-25 00:00:00 , DOI: 10.1021/acssynbio.8b00110 Christopher Sauer 1, 2 , Emiel Ver Loren van Themaat 2 , Leonie G M Boender 2 , Daphne Groothuis 2 , Rita Cruz 1, 2 , Leendert W Hamoen 1, 3 , Colin R Harwood 1 , Tjeerd van Rij 2
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-06-25 00:00:00 , DOI: 10.1021/acssynbio.8b00110 Christopher Sauer 1, 2 , Emiel Ver Loren van Themaat 2 , Leonie G M Boender 2 , Daphne Groothuis 2 , Rita Cruz 1, 2 , Leendert W Hamoen 1, 3 , Colin R Harwood 1 , Tjeerd van Rij 2
Affiliation
|
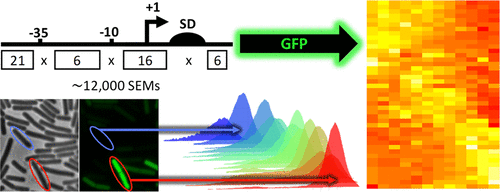
Increasing protein expression levels is a key step in the commercial production of enzymes. Predicting promoter activity and translation initiation efficiency based solely on consensus sequences have so far met with mixed results. Here, we addressed this challenge using a “brute-force” approach by designing and synthesizing a large combinatorial library comprising ∼12 000 unique synthetic expression modules (SEMs) for Bacillus subtilis. Using GFP fluorescence as a reporter of gene expression, we obtained a dynamic expression range that spanned 5 orders of magnitude, as well as a maximal 13-fold increase in expression compared with that of the already strong veg expression module. Analyses of the synthetic modules indicated that sequences at the 5′-end of the mRNA were the most important contributing factor to the differences in expression levels, presumably by preventing formation of strong secondary mRNA structures that affect translation initiation. When the gfp coding region was replaced by the coding region of the xynA gene, encoding the industrially relevant B. subtilis xylanase enzyme, only a 3-fold improvement in xylanase production was observed. Moreover, the correlation between GFP and xylanase expression levels was weak. This suggests that the differences in expression levels between the gfp and xynA constructs were due to differences in 5′-end mRNA folding and consequential differences in the rates of translation initiation. Our data show that the use of large libraries of SEMs, in combination with high-throughput technologies, is a powerful approach to improve the production of a specific protein, but that the outcome cannot necessarily be extrapolated to other proteins.
中文翻译:

探索枯草芽孢杆菌合成表达模块的非保守序列空间
提高蛋白质表达水平是酶商业化生产的关键步骤。迄今为止,仅基于共有序列预测启动子活性和翻译起始效率的结果喜忧参半。在这里,我们通过设计和合成一个包含约 12000 个独特的枯草芽孢杆菌合成表达模块 (SEM) 的大型组合文库,使用“蛮力”方法解决了这一挑战。使用 GFP 荧光作为基因表达的报告者,我们获得了跨越 5 个数量级的动态表达范围,与已经很强大的蔬菜相比,表达量最大增加了 13 倍表达模块。对合成模块的分析表明,mRNA 5' 端的序列是导致表达水平差异的最重要因素,可能是通过阻止形成影响翻译起始的强二级 mRNA 结构。当gfp编码区被xynA基因的编码区取代时,编码工业相关的枯草芽孢杆菌木聚糖酶,仅观察到木聚糖酶产量提高了 3 倍。此外,GFP 和木聚糖酶表达水平之间的相关性较弱。这表明gfp和xynA之间的表达水平差异构建体是由于 5' 端 mRNA 折叠的差异和翻译起始率的相应差异。我们的数据表明,使用大型 SEM 文库与高通量技术相结合,是提高特定蛋白质产量的有效方法,但结果不一定能外推到其他蛋白质。
更新日期:2018-06-25
中文翻译:

探索枯草芽孢杆菌合成表达模块的非保守序列空间
提高蛋白质表达水平是酶商业化生产的关键步骤。迄今为止,仅基于共有序列预测启动子活性和翻译起始效率的结果喜忧参半。在这里,我们通过设计和合成一个包含约 12000 个独特的枯草芽孢杆菌合成表达模块 (SEM) 的大型组合文库,使用“蛮力”方法解决了这一挑战。使用 GFP 荧光作为基因表达的报告者,我们获得了跨越 5 个数量级的动态表达范围,与已经很强大的蔬菜相比,表达量最大增加了 13 倍表达模块。对合成模块的分析表明,mRNA 5' 端的序列是导致表达水平差异的最重要因素,可能是通过阻止形成影响翻译起始的强二级 mRNA 结构。当gfp编码区被xynA基因的编码区取代时,编码工业相关的枯草芽孢杆菌木聚糖酶,仅观察到木聚糖酶产量提高了 3 倍。此外,GFP 和木聚糖酶表达水平之间的相关性较弱。这表明gfp和xynA之间的表达水平差异构建体是由于 5' 端 mRNA 折叠的差异和翻译起始率的相应差异。我们的数据表明,使用大型 SEM 文库与高通量技术相结合,是提高特定蛋白质产量的有效方法,但结果不一定能外推到其他蛋白质。











































 京公网安备 11010802027423号
京公网安备 11010802027423号