Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A distinct abundant group of microbial rhodopsins discovered using functional metagenomics
Nature ( IF 50.5 ) Pub Date : 2018-06-01 , DOI: 10.1038/s41586-018-0225-9 Alina Pushkarev 1 , Keiichi Inoue 2, 3, 4, 5 , Shirley Larom 1 , José Flores-Uribe 1 , Manish Singh 2 , Masae Konno 2 , Sahoko Tomida 2 , Shota Ito 2 , Ryoko Nakamura 2 , Satoshi P Tsunoda 2, 5 , Alon Philosof 1 , Itai Sharon 6, 7 , Natalya Yutin 8 , Eugene V Koonin 8 , Hideki Kandori 2, 3 , Oded Béjà 1
Nature ( IF 50.5 ) Pub Date : 2018-06-01 , DOI: 10.1038/s41586-018-0225-9 Alina Pushkarev 1 , Keiichi Inoue 2, 3, 4, 5 , Shirley Larom 1 , José Flores-Uribe 1 , Manish Singh 2 , Masae Konno 2 , Sahoko Tomida 2 , Shota Ito 2 , Ryoko Nakamura 2 , Satoshi P Tsunoda 2, 5 , Alon Philosof 1 , Itai Sharon 6, 7 , Natalya Yutin 8 , Eugene V Koonin 8 , Hideki Kandori 2, 3 , Oded Béjà 1
Affiliation
|
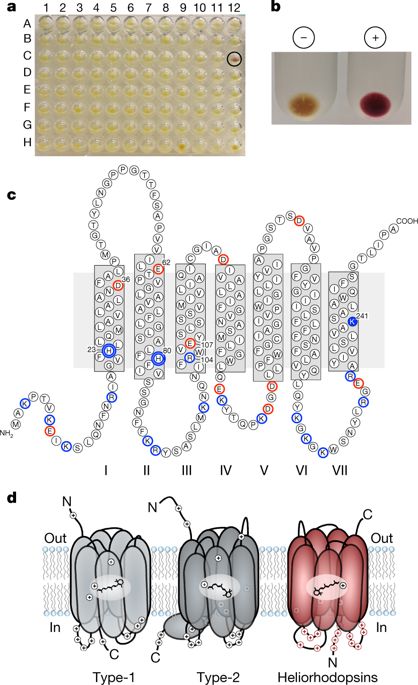
Many organisms capture or sense sunlight using rhodopsin pigments1,2, which are integral membrane proteins that bind retinal chromophores. Rhodopsins comprise two distinct protein families1, type-1 (microbial rhodopsins) and type-2 (animal rhodopsins). The two families share similar topologies and contain seven transmembrane helices that form a pocket in which retinal is linked covalently as a protonated Schiff base to a lysine at the seventh transmembrane helix2,3. Type-1 and type-2 rhodopsins show little or no sequence similarity to each other, as a consequence of extensive divergence from a common ancestor or convergent evolution of similar structures1. Here we report a previously unknown and diverse family of rhodopsins—which we term the heliorhodopsins—that we identified using functional metagenomics and that are distantly related to type-1 rhodopsins. Heliorhodopsins are embedded in the membrane with their N termini facing the cell cytoplasm, an orientation that is opposite to that of type-1 or type-2 rhodopsins. Heliorhodopsins show photocycles that are longer than one second, which is suggestive of light-sensory activity. Heliorhodopsin photocycles accompany retinal isomerization and proton transfer, as in type-1 and type-2 rhodopsins, but protons are never released from the protein, even transiently. Heliorhodopsins are abundant and distributed globally; we detected them in Archaea, Bacteria, Eukarya and their viruses. Our findings reveal a previously unknown family of light-sensing rhodopsins that are widespread in the microbial world.An analysis based on functional metagenomics reveals a previously unknown group of microbial light-sensory rhodopsins that are widespread among a diverse range of microorganisms.
中文翻译:

使用功能宏基因组学发现了一组独特丰富的微生物视紫红质
许多生物体利用视紫红质色素 1,2 捕获或感知阳光,视紫红质色素是结合视网膜发色团的完整膜蛋白。视紫质包含两个不同的蛋白质家族1,1 型(微生物视紫质)和 2 型(动物视紫质)。这两个家族具有相似的拓扑结构,并包含七个跨膜螺旋,形成一个口袋,其中视网膜作为质子化席夫碱与第七个跨膜螺旋处的赖氨酸共价连接2,3。 1 型和 2 型视紫红质彼此之间显示出很少或没有序列相似性,这是由于与共同祖先存在广泛分歧或相似结构趋同进化的结果。在这里,我们报告了一个以前未知的多样化视紫红质家族,我们将其称为日光视紫红质,我们使用功能宏基因组学鉴定了它,并且与 1 型视紫红质有远亲关系。日光视紫红质嵌入细胞膜中,其 N 末端面向细胞质,这一方向与 1 型或 2 型视紫红质相反。日光视紫红质显示出超过一秒的光周期,这表明存在光感应活动。与 1 型和 2 型视紫红质一样,日光视紫红质光循环伴随着视网膜异构化和质子转移,但质子永远不会从蛋白质中释放,即使是短暂的。日光视紫红质丰富且分布于全球;我们在古细菌、细菌、真核生物及其病毒中检测到了它们。我们的研究结果揭示了一个以前未知的光感应视紫红质家族,它们广泛存在于微生物世界中。基于功能宏基因组学的分析揭示了一组以前未知的微生物光感应视紫红质,它们广泛存在于多种微生物中。
更新日期:2018-06-01
中文翻译:

使用功能宏基因组学发现了一组独特丰富的微生物视紫红质
许多生物体利用视紫红质色素 1,2 捕获或感知阳光,视紫红质色素是结合视网膜发色团的完整膜蛋白。视紫质包含两个不同的蛋白质家族1,1 型(微生物视紫质)和 2 型(动物视紫质)。这两个家族具有相似的拓扑结构,并包含七个跨膜螺旋,形成一个口袋,其中视网膜作为质子化席夫碱与第七个跨膜螺旋处的赖氨酸共价连接2,3。 1 型和 2 型视紫红质彼此之间显示出很少或没有序列相似性,这是由于与共同祖先存在广泛分歧或相似结构趋同进化的结果。在这里,我们报告了一个以前未知的多样化视紫红质家族,我们将其称为日光视紫红质,我们使用功能宏基因组学鉴定了它,并且与 1 型视紫红质有远亲关系。日光视紫红质嵌入细胞膜中,其 N 末端面向细胞质,这一方向与 1 型或 2 型视紫红质相反。日光视紫红质显示出超过一秒的光周期,这表明存在光感应活动。与 1 型和 2 型视紫红质一样,日光视紫红质光循环伴随着视网膜异构化和质子转移,但质子永远不会从蛋白质中释放,即使是短暂的。日光视紫红质丰富且分布于全球;我们在古细菌、细菌、真核生物及其病毒中检测到了它们。我们的研究结果揭示了一个以前未知的光感应视紫红质家族,它们广泛存在于微生物世界中。基于功能宏基因组学的分析揭示了一组以前未知的微生物光感应视紫红质,它们广泛存在于多种微生物中。











































 京公网安备 11010802027423号
京公网安备 11010802027423号