当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Coarse-Grained Simulation of Protein-Imprinted Hydrogels
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-07-06 , DOI: 10.1021/acs.jpcb.8b03774 Israel Zadok 1 , Simcha Srebnik 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2018-07-06 , DOI: 10.1021/acs.jpcb.8b03774 Israel Zadok 1 , Simcha Srebnik 1
Affiliation
|
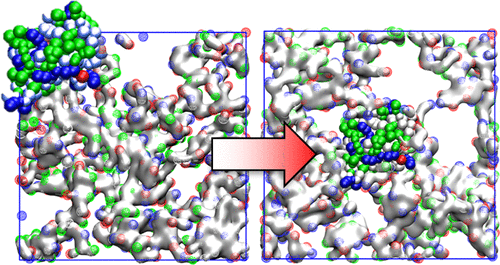
The MARTINI force-field is adapted for acrylic functional monomers and used to simulate protein-imprinted polymers, including complexation in solution, cross-linking reaction, washing, and rebinding of the protein. A globally controlled grand canonical Monte Carlo simulation combined with molecular dynamics is used to achieve proper hydration and swelling of the gel. Two gel formulations are compared, one with only polar functional monomers and one with additional small number of charged monomers. Imprinted hydrogels are evaluated for two proteins, lysozyme, and cytochrome c and chosen due to their similarity in size and isoelectric point, though it is known that their imprinting behavior is very different. Analysis of diffusion of the proteins within the gel shows that their interaction with functional monomers and their diffusion mechanism differ substantially and hence affect their specific and nonspecific interactions with the gel.
中文翻译:

蛋白质印迹水凝胶的粗粒模拟
MARTINI力场适用于丙烯酸功能单体,并用于模拟蛋白质印迹聚合物,包括溶液中的络合,交联反应,洗涤和蛋白质的重新结合。全局控制的大规范蒙特卡洛模拟与分子动力学相结合,可实现凝胶的适当水合作用和溶胀。比较了两种凝胶制剂,一种仅具有极性官能单体,另一种仅具有少量带电单体。印记的水凝胶可评估两种蛋白,溶菌酶和细胞色素c尽管它们的刻印行为非常不同,但由于它们在尺寸和等电点上的相似性而选择它们。蛋白质在凝胶中的扩散分析表明,它们与功能单体的相互作用及其扩散机理存在很大差异,因此会影响它们与凝胶的特异性和非特异性相互作用。
更新日期:2018-07-08
中文翻译:

蛋白质印迹水凝胶的粗粒模拟
MARTINI力场适用于丙烯酸功能单体,并用于模拟蛋白质印迹聚合物,包括溶液中的络合,交联反应,洗涤和蛋白质的重新结合。全局控制的大规范蒙特卡洛模拟与分子动力学相结合,可实现凝胶的适当水合作用和溶胀。比较了两种凝胶制剂,一种仅具有极性官能单体,另一种仅具有少量带电单体。印记的水凝胶可评估两种蛋白,溶菌酶和细胞色素c尽管它们的刻印行为非常不同,但由于它们在尺寸和等电点上的相似性而选择它们。蛋白质在凝胶中的扩散分析表明,它们与功能单体的相互作用及其扩散机理存在很大差异,因此会影响它们与凝胶的特异性和非特异性相互作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号