当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Kinase Substrate Profiling Using a Proteome-wide Serine-Oriented Human Peptide Library
Biochemistry ( IF 2.9 ) Pub Date : 2018-06-19 00:00:00 , DOI: 10.1021/acs.biochem.8b00410 Karl W. Barber 1, 2 , Chad J. Miller 3 , Jay W. Jun 4, 5 , Hua Jane Lou 3 , Benjamin E. Turk 3 , Jesse Rinehart 1, 2, 5
Biochemistry ( IF 2.9 ) Pub Date : 2018-06-19 00:00:00 , DOI: 10.1021/acs.biochem.8b00410 Karl W. Barber 1, 2 , Chad J. Miller 3 , Jay W. Jun 4, 5 , Hua Jane Lou 3 , Benjamin E. Turk 3 , Jesse Rinehart 1, 2, 5
Affiliation
|
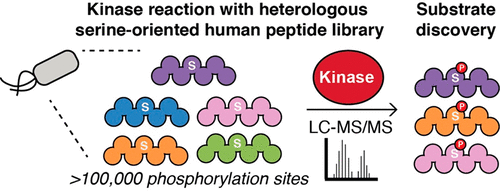
The human proteome encodes >500 protein kinases and hundreds of thousands of potential phosphorylation sites. However, the identification of kinase–substrate pairs remains an active area of research because the relationships between individual kinases and these phosphorylation sites remain largely unknown. Many techniques have been established to discover kinase substrates but are often technically challenging to perform. Moreover, these methods frequently rely on substrate reagent pools that do not reflect human protein sequences or are biased by human cell line protein expression profiles. Here, we describe a new approach called SERIOHL-KILR (serine-oriented human library–kinase library reactions) to profile kinase substrate specificity and to identify candidate substrates for serine kinases. Using a purified library of >100000 serine-oriented human peptides expressed heterologously in Escherichia coli, we perform in vitro kinase reactions to identify phosphorylated human peptide sequences by liquid chromatography and tandem mass spectrometry. We compare our results for protein kinase A to those of a well-established positional scanning peptide library method, certifying that SERIOHL-KILR can identify the same predominant motif elements as traditional techniques. We then interrogate a small panel of cancer-associated PKCβ mutants using our profiling protocol and observe a shift in substrate specificity likely attributable to the loss of key polar contacts between the kinase and its substrates. Overall, we demonstrate that SERIOHL-KILR can rapidly identify candidate kinase substrates that can be directly mapped to human sequences for pathway analysis. Because this technique can be adapted for various kinase studies, we believe that SERIOHL-KILR will have many new victims in the future.
中文翻译:

使用面向蛋白质组的全丝氨酸定向人类肽库进行激酶底物分析
人类蛋白质组编码> 500种蛋白激酶和数十万个潜在的磷酸化位点。但是,激酶-底物对的鉴定仍然是研究的一个活跃领域,因为单个激酶与这些磷酸化位点之间的关系仍然未知。已经建立了许多发现激酶底物的技术,但是通常在技术上难以实施。而且,这些方法经常依赖于不反映人蛋白质序列或受人细胞系蛋白质表达谱偏倚的底物试剂库。在这里,我们描述了一种称为SERIOHL-KILR(面向丝氨酸的人文库-激酶文库反应)的新方法,用于分析激酶底物特异性并鉴定丝氨酸激酶的候选底物。使用>的纯化文库大肠杆菌,我们在体外进行激酶反应,通过液相色谱和串联质谱鉴定磷酸化的人源肽序列。我们将蛋白激酶A的结果与成熟的位置扫描肽库方法的结果进行比较,证明SERIOHL-KILR可以识别与传统技术相同的主要基序元素。然后,我们使用我们的分析方案询问了一小组与癌症相关的PKCβ突变体,并观察到底物特异性的变化,这可能归因于激酶与其底物之间的关键极性接触的丧失。总体而言,我们证明SERIOHL-KILR可以快速识别候选激酶底物,这些底物可以直接定位到人类序列进行途径分析。由于这项技术可适用于各种激酶研究,
更新日期:2018-06-19
中文翻译:

使用面向蛋白质组的全丝氨酸定向人类肽库进行激酶底物分析
人类蛋白质组编码> 500种蛋白激酶和数十万个潜在的磷酸化位点。但是,激酶-底物对的鉴定仍然是研究的一个活跃领域,因为单个激酶与这些磷酸化位点之间的关系仍然未知。已经建立了许多发现激酶底物的技术,但是通常在技术上难以实施。而且,这些方法经常依赖于不反映人蛋白质序列或受人细胞系蛋白质表达谱偏倚的底物试剂库。在这里,我们描述了一种称为SERIOHL-KILR(面向丝氨酸的人文库-激酶文库反应)的新方法,用于分析激酶底物特异性并鉴定丝氨酸激酶的候选底物。使用>的纯化文库大肠杆菌,我们在体外进行激酶反应,通过液相色谱和串联质谱鉴定磷酸化的人源肽序列。我们将蛋白激酶A的结果与成熟的位置扫描肽库方法的结果进行比较,证明SERIOHL-KILR可以识别与传统技术相同的主要基序元素。然后,我们使用我们的分析方案询问了一小组与癌症相关的PKCβ突变体,并观察到底物特异性的变化,这可能归因于激酶与其底物之间的关键极性接触的丧失。总体而言,我们证明SERIOHL-KILR可以快速识别候选激酶底物,这些底物可以直接定位到人类序列进行途径分析。由于这项技术可适用于各种激酶研究,











































 京公网安备 11010802027423号
京公网安备 11010802027423号