当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
MixMD Probeview: Robust Binding Site Prediction from Cosolvent Simulations
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-06-15 00:00:00 , DOI: 10.1021/acs.jcim.8b00265 Sarah E. Graham , Noah Leja , Heather A. Carlson
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-06-15 00:00:00 , DOI: 10.1021/acs.jcim.8b00265 Sarah E. Graham , Noah Leja , Heather A. Carlson
|
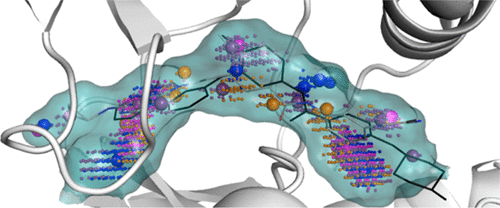
Mixed-solvent molecular dynamics (MixMD) is a cosolvent simulation technique for identifying binding hotspots and specific favorable interactions on a protein’s surface. MixMD studies have the ability to identify these biologically relevant sites by examining the occupancy of the cosolvent over the course of the simulation. However, previous MixMD analysis required a great deal of manual inspection to identify relevant sites. To address this limitation, we have developed MixMD Probeview as a plugin for the freely available, open-source version of the molecular visualization program PyMOL. MixMD Probeview incorporates two analysis procedures: (1) identifying and ranking whole binding sites and (2) identifying and ranking local maxima for each probe type. These functionalities were validated using four common benchmark proteins, including two with both active and allosteric sites. In addition, three different cosolvent procedures were compared to examine the impact of including more than one cosolvent in the simulations. For all systems tested, MixMD Probeview successfully identified known active and allosteric sites based on the total occupancy of neutral probe molecules. As an easy-to-use PyMOL plugin, we expect that MixMD Probeview will facilitate identification and analysis of binding sites from cosolvent simulations performed on a wide range of systems.
中文翻译:

MixMD Probeview:助溶剂模拟中可靠的结合位点预测
混合溶剂分子动力学(MixMD)是一种共溶剂模拟技术,用于识别蛋白质表面上的结合热点和特定的有利相互作用。通过在模拟过程中检查助溶剂的占用情况,MixMD研究具有识别这些生物学相关位点的能力。但是,以前的MixMD分析需要大量的人工检查才能确定相关的位置。为了解决这个限制,我们开发了MixMD Probeview作为分子可视化程序PyMOL的免费开放源代码版本的插件。MixMD Probeview包含两个分析程序:(1)识别和排列整个结合位点,以及(2)识别和排列每种探针类型的局部最大值。这些功能已使用四种常见的基准蛋白进行了验证,包括两个具有活跃和变构位点的位置。另外,比较了三种不同的助溶剂程序,以检查在模拟中包含一种以上助溶剂的影响。对于所有测试的系统,MixMD Probeview根据中性探针分子的总占有率成功确定了已知的活性和变构位点。作为一个易于使用的PyMOL插件,我们希望MixMD Probeview将有助于在多种系统上进行的助溶剂模拟中识别和分析结合位点。MixMD Probeview根据中性探针分子的总占有率成功鉴定出了已知的活性和变构位点。作为一个易于使用的PyMOL插件,我们希望MixMD Probeview将有助于在多种系统上进行的助溶剂模拟中识别和分析结合位点。MixMD Probeview根据中性探针分子的总占有率成功鉴定出了已知的活性和变构位点。作为一个易于使用的PyMOL插件,我们希望MixMD Probeview将有助于在多种系统上进行的助溶剂模拟中识别和分析结合位点。
更新日期:2018-06-15
中文翻译:

MixMD Probeview:助溶剂模拟中可靠的结合位点预测
混合溶剂分子动力学(MixMD)是一种共溶剂模拟技术,用于识别蛋白质表面上的结合热点和特定的有利相互作用。通过在模拟过程中检查助溶剂的占用情况,MixMD研究具有识别这些生物学相关位点的能力。但是,以前的MixMD分析需要大量的人工检查才能确定相关的位置。为了解决这个限制,我们开发了MixMD Probeview作为分子可视化程序PyMOL的免费开放源代码版本的插件。MixMD Probeview包含两个分析程序:(1)识别和排列整个结合位点,以及(2)识别和排列每种探针类型的局部最大值。这些功能已使用四种常见的基准蛋白进行了验证,包括两个具有活跃和变构位点的位置。另外,比较了三种不同的助溶剂程序,以检查在模拟中包含一种以上助溶剂的影响。对于所有测试的系统,MixMD Probeview根据中性探针分子的总占有率成功确定了已知的活性和变构位点。作为一个易于使用的PyMOL插件,我们希望MixMD Probeview将有助于在多种系统上进行的助溶剂模拟中识别和分析结合位点。MixMD Probeview根据中性探针分子的总占有率成功鉴定出了已知的活性和变构位点。作为一个易于使用的PyMOL插件,我们希望MixMD Probeview将有助于在多种系统上进行的助溶剂模拟中识别和分析结合位点。MixMD Probeview根据中性探针分子的总占有率成功鉴定出了已知的活性和变构位点。作为一个易于使用的PyMOL插件,我们希望MixMD Probeview将有助于在多种系统上进行的助溶剂模拟中识别和分析结合位点。



























 京公网安备 11010802027423号
京公网安备 11010802027423号