当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Coupling Green Fluorescent Protein Expression with Chemical Modification to Probe Functionally Relevant Riboswitch Conformations in Live Bacteria
Biochemistry ( IF 2.9 ) Pub Date : 2018-06-13 00:00:00 , DOI: 10.1021/acs.biochem.8b00316 Debapratim Dutta 1 , Ivan A. Belashov 1 , Joseph E. Wedekind 1
Biochemistry ( IF 2.9 ) Pub Date : 2018-06-13 00:00:00 , DOI: 10.1021/acs.biochem.8b00316 Debapratim Dutta 1 , Ivan A. Belashov 1 , Joseph E. Wedekind 1
Affiliation
|
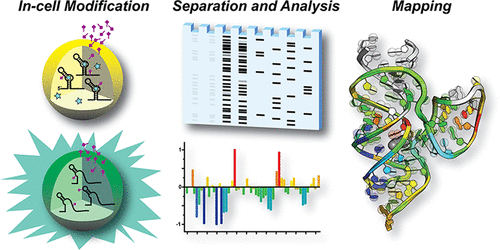
Noncoding RNAs engage in numerous biological activities including gene regulation. To fully understand RNA function it is necessary to probe biologically relevant conformations in living cells. To address this challenge, we coupled RNA-mediated regulation of the green fluorescent protein (GFP)uv-reporter gene to icSHAPE (in cell Selective 2′-Hydroxyl Acylation analyzed by Primer Extension). Our transcript-specific approach provides sensitive, fluorescence-based readout of the regulatory-RNA status as a means to coordinate chemical modification experiments. We chose a plasmid-based reporter compatible with Escherichia coli to allow use of knockout strains that eliminate endogenous effector biosynthesis. The approach was piloted using the Lactobacillus rhamnosus (Lrh) preQ1-II riboswitch, which senses the pyrrolopyrimidine metabolite preQ1. Using an E. coli ΔqueF strain incapable of preQ1 anabolism, the Lrh riboswitch yielded nearly one log unit of GFPuv-gene repression resulting from exogenously added preQ1. We then subjected cells in gene “on” and “off” states to icSHAPE. The resulting differential analysis indicated reduction in Lrh riboswitch flexibility in the P3 helix of the pseudoknot, which comprises the ribosome-binding site (RBS) paired with the anti-RBS. Such expression platform modulation was not observed by in vitro chemical probing and demonstrates that the crowded cellular environment does not preclude detection of compact and loose RNA-regulatory conformations. Here we describe the design, methods, interpretation, and caveats of Reporter Coupled (ReCo) icSHAPE. We also describe mapping of the differential ReCo-icSHAPE results onto the Lrh riboswitch-preQ1 cocrystal structure. The approach should be readily applicable to functional RNAs triggered by effectors or environmental variations.
中文翻译:

绿色荧光蛋白表达与化学修饰的耦合,以探测功能相关的Riboswitch构象在活细菌中。
非编码RNA参与许多生物活动,包括基因调控。要完全了解RNA功能,有必要探查活细胞中生物学相关的构象。为了应对这一挑战,我们耦合在所述绿色荧光蛋白的RNA介导的调控(GFP)UV-报告基因icSHAPE(我Ñ ç ELL小号择2'- ħ ydroxyl甲cylation通过分析P绞刀,决定ë xtension)。我们的转录本特异性方法可提供敏感的,基于荧光的调节性RNA状态读数,作为协调化学修饰实验的一种手段。我们选择了与大肠杆菌兼容的基于质粒的报告基因允许使用消除内源性效应子生物合成的敲除菌株。该方法使用鼠李糖乳杆菌(Lrh)preQ 1 -II核糖开关进行试点,该传感器可检测吡咯并嘧啶的代谢产物preQ 1。使用大肠杆菌Δ queF PREQ的应变无法1合成代谢中,LRH核糖开关产生从外源添加的PREQ所得GFPuv基因抑制近一个日志单元1。然后,我们对处于“打开”和“关闭”状态的基因的细胞进行了icSHAPE处理。产生的差异分析表明Lrh降低假结P3螺旋中的核糖开关柔性,它包括与抗RBS配对的核糖体结合位点(RBS)。体外化学探测未观察到这种表达平台调节,证明了拥挤的细胞环境并不排除检测紧实和松散的RNA调节构象。在这里,我们描述设计,方法,解释和警告的重新搬运工有限公司upled(RECO)icSHAPE。我们还描述了将差分ReCo-icSHAPE结果映射到Lrh核糖开关-preQ 1共晶体结构上的情况。该方法应易于适用于效应子或环境变化触发的功能性RNA。
更新日期:2018-06-13
中文翻译:

绿色荧光蛋白表达与化学修饰的耦合,以探测功能相关的Riboswitch构象在活细菌中。
非编码RNA参与许多生物活动,包括基因调控。要完全了解RNA功能,有必要探查活细胞中生物学相关的构象。为了应对这一挑战,我们耦合在所述绿色荧光蛋白的RNA介导的调控(GFP)UV-报告基因icSHAPE(我Ñ ç ELL小号择2'- ħ ydroxyl甲cylation通过分析P绞刀,决定ë xtension)。我们的转录本特异性方法可提供敏感的,基于荧光的调节性RNA状态读数,作为协调化学修饰实验的一种手段。我们选择了与大肠杆菌兼容的基于质粒的报告基因允许使用消除内源性效应子生物合成的敲除菌株。该方法使用鼠李糖乳杆菌(Lrh)preQ 1 -II核糖开关进行试点,该传感器可检测吡咯并嘧啶的代谢产物preQ 1。使用大肠杆菌Δ queF PREQ的应变无法1合成代谢中,LRH核糖开关产生从外源添加的PREQ所得GFPuv基因抑制近一个日志单元1。然后,我们对处于“打开”和“关闭”状态的基因的细胞进行了icSHAPE处理。产生的差异分析表明Lrh降低假结P3螺旋中的核糖开关柔性,它包括与抗RBS配对的核糖体结合位点(RBS)。体外化学探测未观察到这种表达平台调节,证明了拥挤的细胞环境并不排除检测紧实和松散的RNA调节构象。在这里,我们描述设计,方法,解释和警告的重新搬运工有限公司upled(RECO)icSHAPE。我们还描述了将差分ReCo-icSHAPE结果映射到Lrh核糖开关-preQ 1共晶体结构上的情况。该方法应易于适用于效应子或环境变化触发的功能性RNA。











































 京公网安备 11010802027423号
京公网安备 11010802027423号