Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Quantitative phosphoproteomic analysis of the molecular substrates of sleep need
Nature ( IF 64.8 ) Pub Date : 2018-06-01 , DOI: 10.1038/s41586-018-0218-8 Zhiqiang Wang 1 , Jing Ma 1 , Chika Miyoshi 1 , Yuxin Li 2 , Makito Sato 1 , Yukino Ogawa 1 , Tingting Lou 1 , Chengyuan Ma 3 , Xue Gao 3 , Chiyu Lee 1 , Tomoyuki Fujiyama 1 , Xiaojie Yang 1 , Shuang Zhou 3 , Noriko Hotta-Hirashima 1 , Daniela Klewe-Nebenius 1 , Aya Ikkyu 1 , Miyo Kakizaki 1 , Satomi Kanno 1 , Liqin Cao 1 , Satoru Takahashi 4 , Junmin Peng 2 , Yonghao Yu 5 , Hiromasa Funato 1, 6 , Masashi Yanagisawa 1, 7, 8 , Qinghua Liu 1, 3, 9, 10
Nature ( IF 64.8 ) Pub Date : 2018-06-01 , DOI: 10.1038/s41586-018-0218-8 Zhiqiang Wang 1 , Jing Ma 1 , Chika Miyoshi 1 , Yuxin Li 2 , Makito Sato 1 , Yukino Ogawa 1 , Tingting Lou 1 , Chengyuan Ma 3 , Xue Gao 3 , Chiyu Lee 1 , Tomoyuki Fujiyama 1 , Xiaojie Yang 1 , Shuang Zhou 3 , Noriko Hotta-Hirashima 1 , Daniela Klewe-Nebenius 1 , Aya Ikkyu 1 , Miyo Kakizaki 1 , Satomi Kanno 1 , Liqin Cao 1 , Satoru Takahashi 4 , Junmin Peng 2 , Yonghao Yu 5 , Hiromasa Funato 1, 6 , Masashi Yanagisawa 1, 7, 8 , Qinghua Liu 1, 3, 9, 10
Affiliation
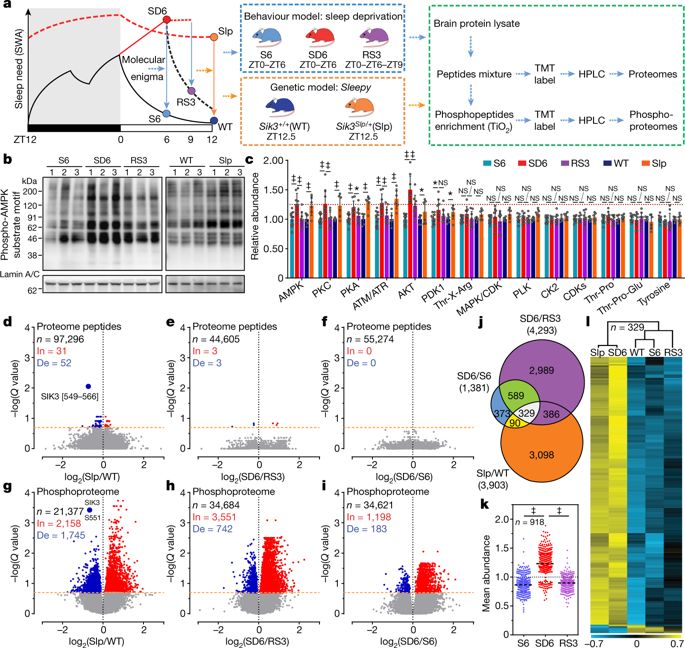
|
Sleep and wake have global effects on brain physiology, from molecular changes1–4 and neuronal activities to synaptic plasticity3–7. Sleep–wake homeostasis is maintained by the generation of a sleep need that accumulates during waking and dissipates during sleep8–11. Here we investigate the molecular basis of sleep need using quantitative phosphoproteomic analysis of the sleep-deprived and Sleepy mouse models of increased sleep need. Sleep deprivation induces cumulative phosphorylation of the brain proteome, which dissipates during sleep. Sleepy mice, owing to a gain-of-function mutation in the Sik3 gene12, have a constitutively high sleep need despite increased sleep amount. The brain proteome of these mice exhibits hyperphosphorylation, similar to that seen in the brain of sleep-deprived mice. Comparison of the two models identifies 80 mostly synaptic sleep-need-index phosphoproteins (SNIPPs), in which phosphorylation states closely parallel changes of sleep need. SLEEPY, the mutant SIK3 protein, preferentially associates with and phosphorylates SNIPPs. Inhibition of SIK3 activity reduces phosphorylation of SNIPPs and slow wave activity during non-rapid-eye-movement sleep, the best known measurable index of sleep need, in both Sleepy mice and sleep-deprived wild-type mice. Our results suggest that phosphorylation of SNIPPs accumulates and dissipates in relation to sleep need, and therefore SNIPP phosphorylation is a molecular signature of sleep need. Whereas waking encodes memories by potentiating synapses, sleep consolidates memories and restores synaptic homeostasis by globally downscaling excitatory synapses4–6. Thus, the phosphorylation–dephosphorylation cycle of SNIPPs may represent a major regulatory mechanism that underlies both synaptic homeostasis and sleep–wake homeostasis.A subset of synaptic proteins are cumulatively phosphorylated during wakefulness and dephosphorylated during sleep, in accordance with sleep need; this may represent a common mechanism underlying regulation of both synaptic homeostasis and sleep–wake homeostasis.
中文翻译:

睡眠需求分子底物的定量磷酸蛋白质组学分析
从分子变化 1-4 和神经元活动到突触可塑性 3-7,睡眠和唤醒对大脑生理有全球影响。睡眠-觉醒动态平衡是通过睡眠需求的产生来维持的,这种需求在清醒期间积累并在睡眠期间消散8-11。在这里,我们使用睡眠剥夺和睡眠需求增加的困倦小鼠模型的定量磷酸蛋白质组学分析来研究睡眠需求的分子基础。睡眠剥夺会导致大脑蛋白质组的累积磷酸化,这种磷酸化会在睡眠期间消散。由于 Sik3 基因 12 的功能获得性突变,嗜睡的小鼠尽管睡眠量增加,但其睡眠需求一直很高。这些小鼠的大脑蛋白质组表现出过度磷酸化,类似于在睡眠剥夺小鼠的大脑中看到的。这两种模型的比较确定了 80 种主要是突触睡眠需要指数磷蛋白 (SNIPP),其中磷酸化状态与睡眠需要的变化密切相关。SLEEPY,突变体 SIK3 蛋白,优先与 SNIPPs 结合并使其磷酸化。抑制 SIK3 活性可降低嗜睡小鼠和睡眠剥夺的野生型小鼠在非快速眼动睡眠期间 SNIPP 的磷酸化和慢波活动,这是最着名的睡眠需求可测量指标。我们的研究结果表明,SNIPP 的磷酸化会随着睡眠需要而积累和消散,因此 SNIPP 磷酸化是睡眠需要的分子特征。清醒通过增强突触对记忆进行编码,而睡眠通过全局缩小兴奋性突触 4-6 来巩固记忆并恢复突触稳态。因此,SNIPPs 的磷酸化-去磷酸化循环可能代表了一种主要的调节机制,它是突触稳态和睡眠-觉醒稳态的基础。根据睡眠需要,一部分突触蛋白在清醒期间累积磷酸化,在睡眠期间去磷酸化;这可能代表了调节突触稳态和睡眠-觉醒稳态的共同机制。
更新日期:2018-06-01
中文翻译:

睡眠需求分子底物的定量磷酸蛋白质组学分析
从分子变化 1-4 和神经元活动到突触可塑性 3-7,睡眠和唤醒对大脑生理有全球影响。睡眠-觉醒动态平衡是通过睡眠需求的产生来维持的,这种需求在清醒期间积累并在睡眠期间消散8-11。在这里,我们使用睡眠剥夺和睡眠需求增加的困倦小鼠模型的定量磷酸蛋白质组学分析来研究睡眠需求的分子基础。睡眠剥夺会导致大脑蛋白质组的累积磷酸化,这种磷酸化会在睡眠期间消散。由于 Sik3 基因 12 的功能获得性突变,嗜睡的小鼠尽管睡眠量增加,但其睡眠需求一直很高。这些小鼠的大脑蛋白质组表现出过度磷酸化,类似于在睡眠剥夺小鼠的大脑中看到的。这两种模型的比较确定了 80 种主要是突触睡眠需要指数磷蛋白 (SNIPP),其中磷酸化状态与睡眠需要的变化密切相关。SLEEPY,突变体 SIK3 蛋白,优先与 SNIPPs 结合并使其磷酸化。抑制 SIK3 活性可降低嗜睡小鼠和睡眠剥夺的野生型小鼠在非快速眼动睡眠期间 SNIPP 的磷酸化和慢波活动,这是最着名的睡眠需求可测量指标。我们的研究结果表明,SNIPP 的磷酸化会随着睡眠需要而积累和消散,因此 SNIPP 磷酸化是睡眠需要的分子特征。清醒通过增强突触对记忆进行编码,而睡眠通过全局缩小兴奋性突触 4-6 来巩固记忆并恢复突触稳态。因此,SNIPPs 的磷酸化-去磷酸化循环可能代表了一种主要的调节机制,它是突触稳态和睡眠-觉醒稳态的基础。根据睡眠需要,一部分突触蛋白在清醒期间累积磷酸化,在睡眠期间去磷酸化;这可能代表了调节突触稳态和睡眠-觉醒稳态的共同机制。



























 京公网安备 11010802027423号
京公网安备 11010802027423号