Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dynamic allostery can drive cold adaptation in enzymes
Nature ( IF 50.5 ) Pub Date : 2018-06-01 , DOI: 10.1038/s41586-018-0183-2 Harry G. Saavedra , James O. Wrabl , Jeremy A. Anderson , Jing Li , Vincent J. Hilser
Nature ( IF 50.5 ) Pub Date : 2018-06-01 , DOI: 10.1038/s41586-018-0183-2 Harry G. Saavedra , James O. Wrabl , Jeremy A. Anderson , Jing Li , Vincent J. Hilser
|
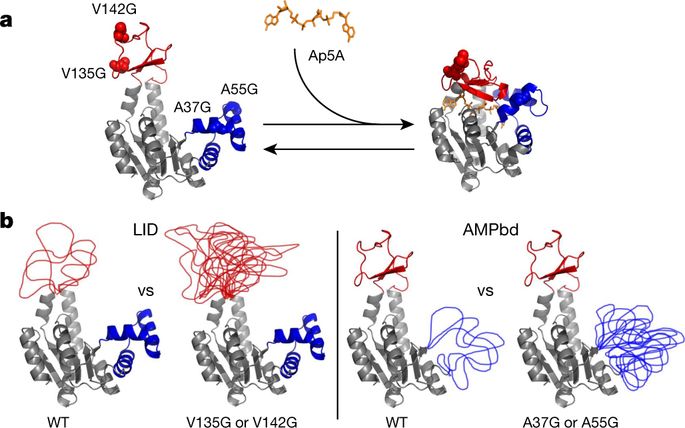
Adaptation of organisms to environmental niches is a hallmark of evolution. One prevalent example is that of thermal adaptation, in which two descendants evolve at different temperature extremes1,2. Underlying the physiological differences between such organisms are changes in enzymes that catalyse essential reactions3, with orthologues from each organism undergoing adaptive mutations that preserve similar catalytic rates at their respective physiological temperatures4,5. The sequence changes responsible for these adaptive differences, however, are often at surface-exposed sites distant from the substrate-binding site, leaving the active site of the enzyme structurally unperturbed6,7. How such changes are allosterically propagated to the active site, to modulate activity, is not known. Here we show that entropy-tuning changes can be engineered into distal sites of Escherichia coli adenylate kinase, allowing us to quantitatively assess the role of dynamics in determining affinity, turnover and the role in driving adaptation. The results not only reveal a dynamics-based allosteric tuning mechanism, but also uncover a spatial separation of the control of key enzymatic parameters. Fluctuations in one mobile domain (the LID) control substrate affinity, whereas dynamic attenuation in the other domain (the AMP-binding domain) affects rate-limiting conformational changes that govern enzyme turnover. Dynamics-based regulation may thus represent an elegant, widespread and previously unrealized evolutionary adaptation mechanism that fine-tunes biological function without altering the ground state structure. Furthermore, because rigid-body conformational changes in both domains were thought to be rate limiting for turnover8,9, these adaptation studies reveal a new model for understanding the relationship between dynamics and turnover in adenylate kinase.By engineering entropy-tuning changes into distal sites of a bacterial adenylate kinase, an allosteric tuning mechanism based on protein dynamics is revealed.
中文翻译:

动态变构可以驱动酶的冷适应
生物体对环境生态位的适应是进化的一个标志。一个普遍的例子是热适应,其中两个后代在不同的极端温度下进化 1,2。这些生物之间的生理差异背后是催化基本反应的酶的变化3,来自每个生物的直系同源物经历适应性突变,在各自的生理温度下保持相似的催化速率4,5。然而,导致这些适应性差异的序列变化通常位于远离底物结合位点的表面暴露位点,使酶的活性位点在结构上不受干扰6,7。尚不清楚这些变化如何变构传播到活性位点以调节活性。在这里,我们表明可以将熵调节变化设计到大肠杆菌腺苷酸激酶的远端位点,使我们能够定量评估动力学在确定亲和力、周转率和驱动适应中的作用。结果不仅揭示了基于动力学的变构调节机制,而且揭示了关键酶参数控制的空间分离。一个移动域(LID)的波动控制底物亲和力,而另一个域(AMP结合域)的动态衰减影响控制酶周转的限速构象变化。因此,基于动力学的调节可能代表了一种优雅、广泛且以前未实现的进化适应机制,可以在不改变基态结构的情况下微调生物功能。此外,
更新日期:2018-06-01
中文翻译:

动态变构可以驱动酶的冷适应
生物体对环境生态位的适应是进化的一个标志。一个普遍的例子是热适应,其中两个后代在不同的极端温度下进化 1,2。这些生物之间的生理差异背后是催化基本反应的酶的变化3,来自每个生物的直系同源物经历适应性突变,在各自的生理温度下保持相似的催化速率4,5。然而,导致这些适应性差异的序列变化通常位于远离底物结合位点的表面暴露位点,使酶的活性位点在结构上不受干扰6,7。尚不清楚这些变化如何变构传播到活性位点以调节活性。在这里,我们表明可以将熵调节变化设计到大肠杆菌腺苷酸激酶的远端位点,使我们能够定量评估动力学在确定亲和力、周转率和驱动适应中的作用。结果不仅揭示了基于动力学的变构调节机制,而且揭示了关键酶参数控制的空间分离。一个移动域(LID)的波动控制底物亲和力,而另一个域(AMP结合域)的动态衰减影响控制酶周转的限速构象变化。因此,基于动力学的调节可能代表了一种优雅、广泛且以前未实现的进化适应机制,可以在不改变基态结构的情况下微调生物功能。此外,









































 京公网安备 11010802027423号
京公网安备 11010802027423号