当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Myeloid Cell Leukemia 1 Inhibition: An in Silico Study Using Non-equilibrium Fast Double Annihilation Technology
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-06-06 00:00:00 , DOI: 10.1021/acs.jctc.8b00305 Piero Procacci 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-06-06 00:00:00 , DOI: 10.1021/acs.jctc.8b00305 Piero Procacci 1
Affiliation
|
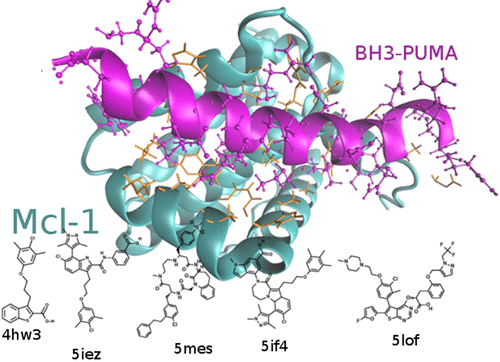
In this work, we compute, by means of a non-equilibrium alchemical technique (fast switching double annihilation methods, FSDAMs), the dissociation free energy for five recently discovered micromolar to sub-nanomolar inhibitors of the Myeloid cell leukemia 1 protein, a key regulator in cell survival and death, providing valuable clues in the chemical–physical determinants of Mcl-1 inhibition. Using the same methodology, we attempt the calculation of the dissociation free energy of the BH3 domain from PUMA protein, binding Mcl-1 in the α-helical state. The synthetic ligands have been parametrized using the recently released GAFF2 general force field [http://ambermd.org] by means of the automated assignment tool PrimaDORAC [Procacci, P. J. Chem. Inf. Model. 2017, 57, 1240]. As an important byproduct, this work constitutes hence one of the first and most challenging tryouts for the GAFF2 parameter set. Agreement with experimental measurements is found to be generally satisfactory, validating the GAFF2 parametrization of the ligands and foreseeing a possible role of FSDAM for industrial application in drug discovery.
中文翻译:

髓样细胞白血病1抑制:使用非平衡快速双重An灭技术的计算机模拟研究
在这项工作中,我们通过一种非平衡的炼金术技术(快速切换双an灭方法,FSDAMs),计算了五种最近发现的髓样白血病1蛋白的微摩尔至亚纳摩尔抑制剂的离解自由能,这是一个关键调节细胞存活和死亡,为抑制Mcl-1的化学-物理决定因素提供了有价值的线索。使用相同的方法,我们尝试计算BH3域与PUMA蛋白的结合自由能,该结合能以α螺旋状态结合Mcl-1。借助自动分配工具PrimaDORAC []使用最近发布的GAFF2通用力场[http://ambermd.org]对合成的配体进行了参数化。Procacci,P。 J.化学。Inf。模型。 2017年, 57,1240]。作为重要的副产品,这项工作因此成为GAFF2参数集的第一个也是最具挑战性的尝试之一。发现与实验测量结果的一致性通常是令人满意的,这验证了配体的GAFF2参数化并预见了FSDAM在药物发现中的工业应用的可能作用。
更新日期:2018-06-06
中文翻译:

髓样细胞白血病1抑制:使用非平衡快速双重An灭技术的计算机模拟研究
在这项工作中,我们通过一种非平衡的炼金术技术(快速切换双an灭方法,FSDAMs),计算了五种最近发现的髓样白血病1蛋白的微摩尔至亚纳摩尔抑制剂的离解自由能,这是一个关键调节细胞存活和死亡,为抑制Mcl-1的化学-物理决定因素提供了有价值的线索。使用相同的方法,我们尝试计算BH3域与PUMA蛋白的结合自由能,该结合能以α螺旋状态结合Mcl-1。借助自动分配工具PrimaDORAC []使用最近发布的GAFF2通用力场[http://ambermd.org]对合成的配体进行了参数化。











































 京公网安备 11010802027423号
京公网安备 11010802027423号