当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Sequence-Dependent Three Interaction Site Model for Single- and Double-Stranded DNA
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-06-05 00:00:00 , DOI: 10.1021/acs.jctc.8b00091 Debayan Chakraborty 1 , Naoto Hori 1 , D Thirumalai 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-06-05 00:00:00 , DOI: 10.1021/acs.jctc.8b00091 Debayan Chakraborty 1 , Naoto Hori 1 , D Thirumalai 1
Affiliation
|
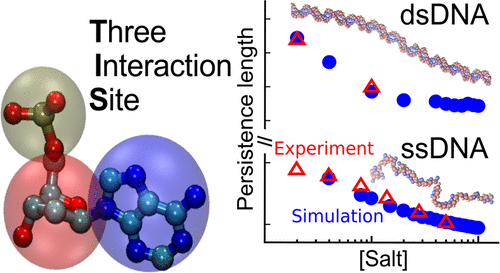
We develop a robust coarse-grained model for single- and double-stranded DNA by representing each nucleotide by three interaction sites (TIS) located at the centers of mass of sugar, phosphate, and base. The resulting TIS model includes base-stacking, hydrogen bond, and electrostatic interactions as well as bond-stretching and bond angle potentials that account for the polymeric nature of DNA. The choices of force constants for stretching and the bending potentials were guided by a Boltzmann inversion procedure using a large representative set of DNA structures extracted from the Protein Data Bank. Some of the parameters in the stacking interactions were calculated using a learning procedure, which ensured that the experimentally measured melting temperatures of dimers are faithfully reproduced. Without any further adjustments, the calculations based on the TIS model reproduce the experimentally measured salt and sequence-dependence of the size of single-stranded DNA (ssDNA), as well as the persistence lengths of poly(dA) and poly(dT) chains. Interestingly, upon application of mechanical force, the extension of poly(dA) exhibits a plateau, which we trace to the formation of stacked helical domains. In contrast, the force–extension curve (FEC) of poly(dT) is entropic in origin and could be described by a standard polymer model. We also show that the persistence length of double-stranded DNA, formed from two complementary ssDNAs, is consistent with the prediction based on the worm-like chain. The persistence length, which decreases with increasing salt concentration, is in accord with the Odijk-Skolnick-Fixman theory intended for stiff polyelectrolyte chains near the rod limit. Our model predicts the melting temperatures of DNA hairpins with excellent accuracy, and we are able to recover the experimentally known sequence-specific trends. The range of applications, which did not require adjusting any parameter after the initial construction based solely on PDB structures and melting profiles of dimers, attests to the transferability and robustness of the TIS model for ssDNA and dsDNA.
中文翻译:

单链和双链 DNA 的序列依赖性三相互作用位点模型
我们通过位于糖、磷酸盐和碱基质量中心的三个相互作用位点 (TIS) 代表每个核苷酸,为单链和双链 DNA 开发了一个强大的粗粒度模型。由此产生的 TIS 模型包括碱基堆积、氢键和静电相互作用,以及解释 DNA 聚合性质的键拉伸和键角势。拉伸力常数和弯曲势的选择是通过玻尔兹曼反演程序来指导的,该程序使用从蛋白质数据库中提取的大量代表性 DNA 结构集。堆叠相互作用中的一些参数是使用学习程序计算的,这确保了实验测量的二聚体熔化温度得到忠实再现。无需任何进一步调整,基于 TIS 模型的计算即可再现实验测量的单链 DNA (ssDNA) 大小的盐和序列依赖性,以及聚 (dA) 和聚 (dT) 链的持久长度。有趣的是,在施加机械力时,聚(dA)的延伸表现出一个平台期,我们将其追溯到堆叠螺旋域的形成。相比之下,poly(dT) 的力-延伸曲线 (FEC) 本质上是熵,可以用标准聚合物模型来描述。我们还表明,由两条互补的 ssDNA 形成的双链 DNA 的持久长度与基于蠕虫状链的预测一致。持久长度随着盐浓度的增加而减小,符合 Odijk-Skolnick-Fixman 理论,该理论适用于接近杆极限的刚性聚电解质链。我们的模型以极高的准确性预测 DNA 发夹的解链温度,并且我们能够恢复实验上已知的序列特异性趋势。仅基于 PDB 结构和二聚体熔解曲线进行初始构建后不需要调整任何参数的应用范围证明了 ssDNA 和 dsDNA 的 TIS 模型的可转移性和稳健性。
更新日期:2018-06-05
中文翻译:

单链和双链 DNA 的序列依赖性三相互作用位点模型
我们通过位于糖、磷酸盐和碱基质量中心的三个相互作用位点 (TIS) 代表每个核苷酸,为单链和双链 DNA 开发了一个强大的粗粒度模型。由此产生的 TIS 模型包括碱基堆积、氢键和静电相互作用,以及解释 DNA 聚合性质的键拉伸和键角势。拉伸力常数和弯曲势的选择是通过玻尔兹曼反演程序来指导的,该程序使用从蛋白质数据库中提取的大量代表性 DNA 结构集。堆叠相互作用中的一些参数是使用学习程序计算的,这确保了实验测量的二聚体熔化温度得到忠实再现。无需任何进一步调整,基于 TIS 模型的计算即可再现实验测量的单链 DNA (ssDNA) 大小的盐和序列依赖性,以及聚 (dA) 和聚 (dT) 链的持久长度。有趣的是,在施加机械力时,聚(dA)的延伸表现出一个平台期,我们将其追溯到堆叠螺旋域的形成。相比之下,poly(dT) 的力-延伸曲线 (FEC) 本质上是熵,可以用标准聚合物模型来描述。我们还表明,由两条互补的 ssDNA 形成的双链 DNA 的持久长度与基于蠕虫状链的预测一致。持久长度随着盐浓度的增加而减小,符合 Odijk-Skolnick-Fixman 理论,该理论适用于接近杆极限的刚性聚电解质链。我们的模型以极高的准确性预测 DNA 发夹的解链温度,并且我们能够恢复实验上已知的序列特异性趋势。仅基于 PDB 结构和二聚体熔解曲线进行初始构建后不需要调整任何参数的应用范围证明了 ssDNA 和 dsDNA 的 TIS 模型的可转移性和稳健性。









































 京公网安备 11010802027423号
京公网安备 11010802027423号