当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Multiplexed CRISPR-Cpf1-Mediated Genome Editing in Clostridium difficile toward the Understanding of Pathogenesis of C. difficile Infection
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-06-04 00:00:00 , DOI: 10.1021/acssynbio.8b00087 Wei Hong , Jie Zhang , Guzhen Cui , Luxin Wang , Yi Wang
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-06-04 00:00:00 , DOI: 10.1021/acssynbio.8b00087 Wei Hong , Jie Zhang , Guzhen Cui , Luxin Wang , Yi Wang
|
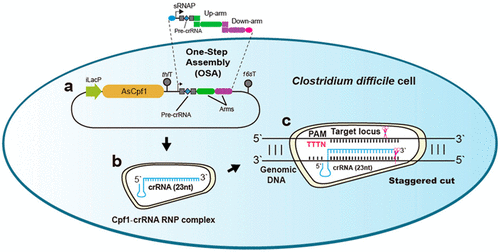
Clostridium difficile is often the primary cause of nosocomial diarrhea, leading to thousands of deaths annually worldwide. The availability of an efficient genome editing tool for C. difficile is essential to understanding its pathogenic mechanism and physiological behavior. Although CRISPR-Cas9 has been extensively employed for genome engineering in various organisms, large gene deletion and multiplex genome editing is still challenging in microorganisms with underdeveloped genetic engineering tools. Here, we describe a streamlined CRISPR-Cpf1-based toolkit to achieve precise deletions of fur, tetM, and ermB1/2 in C. difficile with high efficiencies. All of these genes are relevant to important phenotypes (including iron uptake, antibiotics resistance, and toxin production) as related to the pathogenesis of C. difficile infection (CDI). Furthermore, we were able to delete an extremely large locus of 49.2-kb comprising a phage genome (phiCD630-2) and realized multiplex genome editing in a single conjugation with high efficiencies (simultaneous deletion of cwp66 and tcdA). Our work highlighted the first application of CRISPR-Cpf1 for multiplexed genome editing and extremely large gene deletion in C. difficile, which are both crucial for understanding the pathogenic mechanism of C. difficile and developing strategies to fight against CDI. In addition, for the DNA cloning, we developed a one-step-assembly protocol along with a Python-based algorithm for automatic primer design, shortening the time for plasmid construction to half that of conventional procedures. The approaches we developed herein are easily and broadly applicable to other microorganisms. Our results provide valuable guidance for establishing CRISPR-Cpf1 as a versatile genome engineering tool in prokaryotic cells.
中文翻译:

艰难梭状芽胞杆菌的CRISPR-Cpf1介导的基因组编辑研究对艰难梭菌感染的发病机理的理解
艰难梭菌通常是医院腹泻的主要原因,全世界每年导致数千人死亡。有效的艰难梭菌基因组编辑工具的可用性对于了解其致病机理和生理行为至关重要。尽管CRISPR-Cas9已广泛用于各种生物的基因组工程中,但是对于基因开发不完善的微生物而言,大基因缺失和多重基因组编辑仍对微生物构成挑战。在这里,我们描述了一种简化的基于CRISPR-Cpf1的工具包来实现的精确缺失毛皮,tetM和ermB1 / 2在艰难梭菌效率高。所有这些基因都与重要的表型(包括铁摄取,抗生素抗性和毒素产生)有关,与艰难梭菌感染(CDI)的发病机理有关。此外,我们能够删除一个包含噬菌体基因组(phiCD630-2)的49.2kb超大基因座,并在单个结合中高效地实现了多重基因组编辑(同时删除cwp66和tcdA)。我们的工作着重介绍了CRISPR-Cpf1在艰难梭菌中进行多重基因组编辑和超大基因缺失的首次应用,这对于理解艰难梭菌的致病机理都至关重要。制定打击CDI的策略。此外,对于DNA克隆,我们开发了一步组装协议以及基于Python的自动引物设计算法,从而将质粒构建的时间缩短到传统方法的一半。我们在此开发的方法可轻松,广泛地应用于其他微生物。我们的结果为建立CRISPR-Cpf1作为原核细胞中通用的基因组工程工具提供了宝贵的指导。
更新日期:2018-06-04
中文翻译:

艰难梭状芽胞杆菌的CRISPR-Cpf1介导的基因组编辑研究对艰难梭菌感染的发病机理的理解
艰难梭菌通常是医院腹泻的主要原因,全世界每年导致数千人死亡。有效的艰难梭菌基因组编辑工具的可用性对于了解其致病机理和生理行为至关重要。尽管CRISPR-Cas9已广泛用于各种生物的基因组工程中,但是对于基因开发不完善的微生物而言,大基因缺失和多重基因组编辑仍对微生物构成挑战。在这里,我们描述了一种简化的基于CRISPR-Cpf1的工具包来实现的精确缺失毛皮,tetM和ermB1 / 2在艰难梭菌效率高。所有这些基因都与重要的表型(包括铁摄取,抗生素抗性和毒素产生)有关,与艰难梭菌感染(CDI)的发病机理有关。此外,我们能够删除一个包含噬菌体基因组(phiCD630-2)的49.2kb超大基因座,并在单个结合中高效地实现了多重基因组编辑(同时删除cwp66和tcdA)。我们的工作着重介绍了CRISPR-Cpf1在艰难梭菌中进行多重基因组编辑和超大基因缺失的首次应用,这对于理解艰难梭菌的致病机理都至关重要。制定打击CDI的策略。此外,对于DNA克隆,我们开发了一步组装协议以及基于Python的自动引物设计算法,从而将质粒构建的时间缩短到传统方法的一半。我们在此开发的方法可轻松,广泛地应用于其他微生物。我们的结果为建立CRISPR-Cpf1作为原核细胞中通用的基因组工程工具提供了宝贵的指导。











































 京公网安备 11010802027423号
京公网安备 11010802027423号