Ecotoxicology and Environmental Safety ( IF 6.2 ) Pub Date : 2018-05-21 , DOI: 10.1016/j.ecoenv.2018.05.007 Xing Wang , Xiaoqin Li , Zhenjun Sun
|
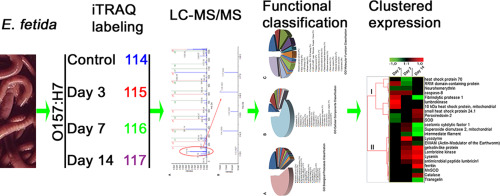
Soil environment contaminated by Escherichia coli O157:H7 which come from the waste of infected animals. Earthworms can live in the pathogens-polluted soil by their innate immunity. How the proteins of earthworms E. fetida will response to E. coli O157:H7-contaminated-soil still unclear? To identify the defense proteins under E. coli O157:H7 stress, we performed a proteomic analysis of earthworm under E. coli O157:H7 exposure through an iTRAQ technology. In total, we found 283 non-redundant proteins, including fibrinolytic protease 1, lombricine kinase, lysozyme, gelsolin, coelomic cytolytic factor-1, antimicrobial peptide lumbricin-l, lysenin, and et al. The proteins participate in metabolic processes, transcription, defense response to bacterium, translation, response to stress, and transport. The study will contribute to understand why earthworm can live in the pathogens-polluted environment.
中文翻译:

基于iTRAQ的定量蛋白质组学分析f对大肠杆菌O157:H7的反应
大肠杆菌O157:H7污染的土壤环境来自被感染动物的废物。can可以通过其固有的免疫力生活在被病原体污染的土壤中。f大肠埃希氏菌的蛋白质如何对受大肠杆菌O157:H7污染的土壤做出反应仍不清楚?为了鉴定大肠杆菌O157:H7胁迫下的防御蛋白,我们在大肠杆菌下进行了worm的蛋白质组学分析通过iTRAQ技术暴露O157:H7。总共,我们发现了283种非冗余蛋白,包括纤溶酶1,lombricine激酶,溶菌酶,凝溶胶蛋白,coelomic cytolytic factor-1,抗菌肽lumbricin-1,lysenin等。这些蛋白质参与代谢过程,转录,对细菌的防御反应,翻译,对应激的反应和运输。这项研究将有助于理解为什么why可以生活在被病原体污染的环境中。









































 京公网安备 11010802027423号
京公网安备 11010802027423号