Bioorganic & Medicinal Chemistry Letters ( IF 2.5 ) Pub Date : 2018-06-01 , DOI: 10.1016/j.bmcl.2018.05.061 Hans Matter , Stefan Güssregen
|
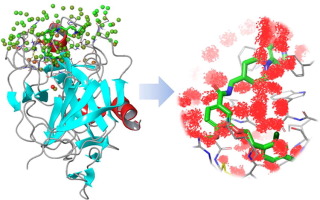
Water is an essential part of protein binding sites and mediates interactions to ligands. Its displacement by ligand parts affects the free binding energy of resulting protein-ligand complexes. Therefore the characterization of solvation properties is important for design. Of particular interest is the propensity of localized water to be favorably displaced by a ligand. This review discusses two popular computational approaches addressing these questions, namely WaterMap based on statistical mechanics analysis of MD simulations and 3D RISM based on integral equation theory of liquids. The theoretical background and recent applications in structure-based design will be presented.
中文翻译:

表征蛋白质-配体复合物中的水合位点,以设计新型配体
水是蛋白质结合位点的重要组成部分,并介导与配体的相互作用。其被配体部分置换会影响所得蛋白质-配体复合物的自由结合能。因此,溶剂化性质的表征对于设计很重要。特别令人感兴趣的是局部水倾向于被配体有利地置换。这篇综述讨论了解决这些问题的两种流行的计算方法,即基于MD模拟的统计力学分析的WaterMap和基于液体积分方程理论的3D RISM。将介绍基于结构的设计的理论背景和最新应用。










































 京公网安备 11010802027423号
京公网安备 11010802027423号