当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Redefining the Protein Kinase Conformational Space with Machine Learning
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2018-05-31 , DOI: 10.1016/j.chembiol.2018.05.002 Peter Man-Un Ung , Rayees Rahman , Avner Schlessinger
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2018-05-31 , DOI: 10.1016/j.chembiol.2018.05.002 Peter Man-Un Ung , Rayees Rahman , Avner Schlessinger
|
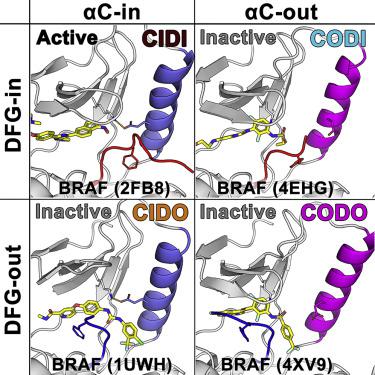
Protein kinases are dynamic, adopting different conformational states that are critical for their catalytic activity. We assess a range of structural features derived from the conserved αC helix and DFG motif to define the conformational space of the catalytic domain of protein kinases. We then constructKinformation, a random forest classifier, to annotate the conformation of 3,708 kinase structures in the PDB. Our classification scheme captures known active and inactive kinase conformations and defines an additional conformational state, thereby refining the current understanding of the kinase conformational space. Furthermore, network analysis of the small molecules recognized by each conformation captures chemical substructures that are associated with each conformation type. Our description of the kinase conformational space is expected to improve modeling of protein kinase structures, as well as guide the development of conformation-specific kinase inhibitors with optimal pharmacological profiles.
中文翻译:

通过机器学习重新定义蛋白质激酶构象空间
蛋白激酶是动态的,采用对其催化活性至关重要的不同构象状态。我们评估了从保守的αC螺旋和DFG基序衍生的一系列结构特征,以定义蛋白激酶催化域的构象空间。然后,我们构造一个随机森林分类器“ Kinformation”,以注释PDB中3,708个激酶结构的构象。我们的分类方案捕获了已知的活性和非活性激酶构象,并定义了其他构象状态,从而完善了对激酶构象空间的当前了解。此外,对每种构象可识别的小分子的网络分析可捕获与每种构象类型相关的化学亚结构。
更新日期:2018-07-20
中文翻译:

通过机器学习重新定义蛋白质激酶构象空间
蛋白激酶是动态的,采用对其催化活性至关重要的不同构象状态。我们评估了从保守的αC螺旋和DFG基序衍生的一系列结构特征,以定义蛋白激酶催化域的构象空间。然后,我们构造一个随机森林分类器“ Kinformation”,以注释PDB中3,708个激酶结构的构象。我们的分类方案捕获了已知的活性和非活性激酶构象,并定义了其他构象状态,从而完善了对激酶构象空间的当前了解。此外,对每种构象可识别的小分子的网络分析可捕获与每种构象类型相关的化学亚结构。











































 京公网安备 11010802027423号
京公网安备 11010802027423号