当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Massively Parallel Implementation of Divide-and-Conquer Jacobi Iterations Using Particle-Mesh Ewald for Force Field Polarization
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-05-30 00:00:00 , DOI: 10.1021/acs.jctc.8b00328 Dominique Nocito 1 , Gregory J. O. Beran 1
Journal of Chemical Theory and Computation ( IF 5.5 ) Pub Date : 2018-05-30 00:00:00 , DOI: 10.1021/acs.jctc.8b00328 Dominique Nocito 1 , Gregory J. O. Beran 1
Affiliation
|
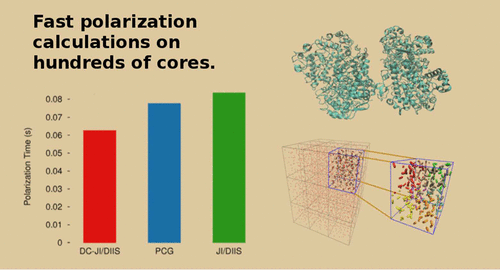
To accelerate the evaluation of the self-consistent polarization in large condensed-phase systems with polarizable force fields, the new divide-and-conquer Jacobi iterations (DC-JI) solver is adapted for periodic boundary conditions with particle-mesh Ewald and implemented in a massively parallel fashion within the Tinker-HP software package. DC-JI captures the mutual polarization of close-range interactions within subclusters of atoms using Cholesky decomposition and couples in the polarization effects between these clusters iteratively. Iterative convergence is accelerated with direct inversion of the iterative subspace (DIIS) extrapolation. Compared to widely used preconditioned conjugate gradient (PCG) or conventional Jacobi iterations (JI/DIIS) algorithms, DC-JI/DIIS solves the polarization equations ∼20–30% faster in protein systems ranging from ∼10,000–175,000 atoms run on hundreds of processor cores. This translates to ∼10–15% speed-ups in the number of nanoseconds of simulation time that can be achieved per day. Not only is DC-JI/DIIS faster than PCG, but it also gives more energetically robust solutions for a given convergence threshold. These improvements make numerically robust polarizable force field simulations more computationally tractable for chemical systems of interest.
中文翻译:

使用粒子网格Ewald进行力场极化的大规模并行实现分而治之Jacobi迭代
为了加快在具有可极化力场的大型凝聚相系统中自洽极化的评估,新的分治式雅可比迭代(DC-JI)求解器适用于粒子网格Ewald的周期性边界条件,并在Tinker-HP软件包中的大规模并行方式。DC-JI使用Cholesky分解捕获原子子簇内近距离相互作用的相互极化,并迭代耦合这些团簇之间的极化效应。通过直接反转迭代子空间(DIIS)外推,可以加速迭代收敛。与广泛使用的预处理共轭梯度(PCG)或常规Jacobi迭代(JI / DIIS)算法相比,在数百个处理器内核上运行的约10,000–175,000个原子的蛋白质系统中,DC-JI / DIIS解决极化方程的速度提高了约20-30%。这相当于每天可以实现的纳秒级仿真速度提高了约10–15%。DC-JI / DIIS不仅比PCG快,而且对于给定的收敛阈值,它还提供了更强大的解决方案。这些改进使数值健壮的可极化力场仿真对于感兴趣的化学系统更易于计算。
更新日期:2018-05-30
中文翻译:

使用粒子网格Ewald进行力场极化的大规模并行实现分而治之Jacobi迭代
为了加快在具有可极化力场的大型凝聚相系统中自洽极化的评估,新的分治式雅可比迭代(DC-JI)求解器适用于粒子网格Ewald的周期性边界条件,并在Tinker-HP软件包中的大规模并行方式。DC-JI使用Cholesky分解捕获原子子簇内近距离相互作用的相互极化,并迭代耦合这些团簇之间的极化效应。通过直接反转迭代子空间(DIIS)外推,可以加速迭代收敛。与广泛使用的预处理共轭梯度(PCG)或常规Jacobi迭代(JI / DIIS)算法相比,在数百个处理器内核上运行的约10,000–175,000个原子的蛋白质系统中,DC-JI / DIIS解决极化方程的速度提高了约20-30%。这相当于每天可以实现的纳秒级仿真速度提高了约10–15%。DC-JI / DIIS不仅比PCG快,而且对于给定的收敛阈值,它还提供了更强大的解决方案。这些改进使数值健壮的可极化力场仿真对于感兴趣的化学系统更易于计算。



























 京公网安备 11010802027423号
京公网安备 11010802027423号