当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structural–elastic determination of the force-dependent transition rate of biomolecules†
Chemical Science ( IF 7.6 ) Pub Date : 2018-05-29 00:00:00 , DOI: 10.1039/c8sc01319e Shiwen Guo 1, 2, 3 , Qingnan Tang 2, 4, 5 , Mingxi Yao 1, 2, 3 , Huijuan You 6, 7, 8, 9, 10 , Shimin Le 2, 4, 5 , Hu Chen 4, 11, 12, 13 , Jie Yan 1, 2, 2, 3, 4
Chemical Science ( IF 7.6 ) Pub Date : 2018-05-29 00:00:00 , DOI: 10.1039/c8sc01319e Shiwen Guo 1, 2, 3 , Qingnan Tang 2, 4, 5 , Mingxi Yao 1, 2, 3 , Huijuan You 6, 7, 8, 9, 10 , Shimin Le 2, 4, 5 , Hu Chen 4, 11, 12, 13 , Jie Yan 1, 2, 2, 3, 4
Affiliation
|
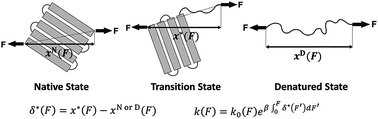
The force-dependent unfolding/refolding of protein domains and ligand-receptor association/dissociation are crucial for mechanosensitive functions, while many aspects of how force affects the transition rate still remain poorly understood. Here, we report a new analytical expression of the force-dependent rate of molecules for transitions overcoming a single barrier. Unlike previous models derived in the framework of Kramers theory that requires a presumed one-dimensional free energy landscape, our model is derived based on the structural–elastic properties of molecules which are not restricted by the shape and dimensionality of the underlying free energy landscape. Importantly, the parameters of this model provide direct information on the structural–elastic features of the molecules between their transition and initial states. We demonstrate the applications of this model by applying it to explain force-dependent transition kinetics for several molecules and predict the structural–elastic properties of the transition states of these molecules.
中文翻译:

生物分子的力依赖转变速率的结构弹性测定†
力依赖的蛋白质结构域的解折叠/折叠以及配体-受体的缔合/解离对于机械敏感功能至关重要,尽管人们对力如何影响转变速率的许多方面仍知之甚少。在这里,我们报告了克服单个障碍的过渡分子的依赖于力的速率的新的分析表达式。不同于先前在Kramers理论框架中得出的,需要一个一维自由能态的模型,我们的模型是基于分子的结构弹性特性而获得的,不受分子的形状和维数的限制。重要的是,该模型的参数提供了有关分子在其过渡态和初始态之间的结构弹性特征的直接信息。
更新日期:2018-05-29
中文翻译:

生物分子的力依赖转变速率的结构弹性测定†
力依赖的蛋白质结构域的解折叠/折叠以及配体-受体的缔合/解离对于机械敏感功能至关重要,尽管人们对力如何影响转变速率的许多方面仍知之甚少。在这里,我们报告了克服单个障碍的过渡分子的依赖于力的速率的新的分析表达式。不同于先前在Kramers理论框架中得出的,需要一个一维自由能态的模型,我们的模型是基于分子的结构弹性特性而获得的,不受分子的形状和维数的限制。重要的是,该模型的参数提供了有关分子在其过渡态和初始态之间的结构弹性特征的直接信息。











































 京公网安备 11010802027423号
京公网安备 11010802027423号