当前位置:
X-MOL 学术
›
Chem. Bio. Drug Des.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
PiViewer: An open‐source tool for automated detection and display of π–π interactions
Chemical Biology & Drug Design ( IF 3.2 ) Pub Date : 2018-06-13 , DOI: 10.1111/cbdd.13340 Ying Liu 1 , Xiang Ao 1 , Qiong Wang 2, 3 , Jianxun Wang 1 , Hu Ge 1, 2
Chemical Biology & Drug Design ( IF 3.2 ) Pub Date : 2018-06-13 , DOI: 10.1111/cbdd.13340 Ying Liu 1 , Xiang Ao 1 , Qiong Wang 2, 3 , Jianxun Wang 1 , Hu Ge 1, 2
Affiliation
|
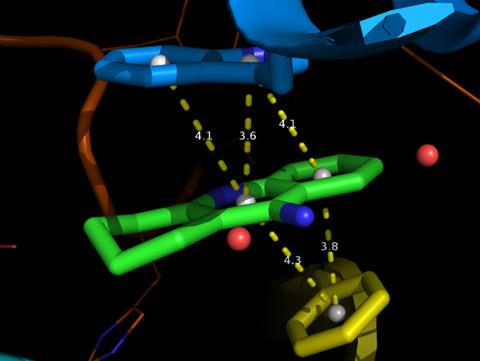
π–π interactions are common and important noncovalent interactions that contribute to biochemical molecular interactions, but the tools for the convenient 3D visualization of π–π interactions are lacking. We have developed an open‐source and easy‐to‐use tool for the automated identification and display of π–π stacking in 3D. It can percept the aromaticity of rings from any selected ligand and the surrounding residues, calculate the distances and dihedral angles between each pair of aromatic ring planes, as well as can highlight the various configurations of π–π interactions in 3D: the sandwich configuration, the T‐shaped configuration, and the parallel‐displaced configuration. In addition, the users can easily adjust or set their own criteria for π–π stacking.
中文翻译:

PiViewer:一个开源工具,用于自动检测和显示π–π相互作用
π – π相互作用是常见且重要的非共价相互作用,可促进生化分子相互作用,但缺乏方便的3D可视化π – π相互作用的工具。我们已经开发了一种开源且易于使用的工具,用于在3D中自动识别和显示π – π堆叠。它可以感知任何选定配体和周围残基的环的芳香性,计算每对芳香环平面之间的距离和二面角,还可以突出显示π – π的各种构型3D交互:三明治配置,T形配置和平行位移配置。此外,用户可以轻松调整或设置自己的π – π堆叠标准。
更新日期:2018-06-13
中文翻译:

PiViewer:一个开源工具,用于自动检测和显示π–π相互作用
π – π相互作用是常见且重要的非共价相互作用,可促进生化分子相互作用,但缺乏方便的3D可视化π – π相互作用的工具。我们已经开发了一种开源且易于使用的工具,用于在3D中自动识别和显示π – π堆叠。它可以感知任何选定配体和周围残基的环的芳香性,计算每对芳香环平面之间的距离和二面角,还可以突出显示π – π的各种构型3D交互:三明治配置,T形配置和平行位移配置。此外,用户可以轻松调整或设置自己的π – π堆叠标准。











































 京公网安备 11010802027423号
京公网安备 11010802027423号