当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dynamic and Structural Modeling of the Specificity in Protein–DNA Interactions Guided by Binding Assay and Structure Data
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-05-28 00:00:00 , DOI: 10.1021/acs.jctc.8b00299 Cheng Tan 1 , Shoji Takada 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-05-28 00:00:00 , DOI: 10.1021/acs.jctc.8b00299 Cheng Tan 1 , Shoji Takada 1
Affiliation
|
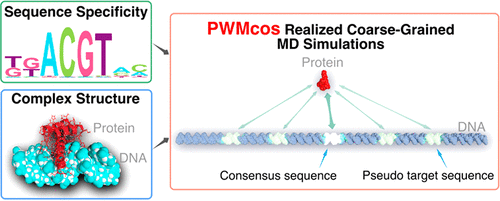
How transcription factors (TFs) recognize their DNA sequences is often investigated complementarily by high-throughput protein binding assays and by structural biology experiments. The former quantifies the specificity of TF binding sites for numerous DNA sequences, often represented as the position-weight-matrix (PWM). The latter provides mechanistic insights into the interactions via the protein–DNA complex structures. However, these two types of data are not readily integrated. Here, we propose and test a new modeling method that incorporates the PWM with complex structure data. On the basis of pretuned coarse-grained models for proteins and DNAs, we model the specific protein–DNA interactions, PWMcos, in terms of an orientation-dependent potential function, which enables us to perform molecular dynamics simulations at unprecedentedly large scales. We show that the PWMcos model reproduces subtle specificity in the protein–DNA recognition. During the target search in genomic sequences, TF moves on highly rugged landscapes and occasionally flips on DNA depending on the sequence. The TATA-binding protein exhibits two remarkably distinct binding modes, of which frequencies differ between TATA-containing and TATA-less promoters. The PWMcos is general and can be applied to any protein–DNA interactions given their PWMs and complex structure data are available.
中文翻译:

结合测定和结构数据指导的蛋白质-DNA相互作用特异性的动态和结构模型
转录因子(TFs)如何识别其DNA序列通常通过高通量蛋白质结合测定和结构生物学实验进行补充研究。前者可量化TF结合位点对许多DNA序列的特异性,这些序列通常表示为位置权重矩阵(PWM)。后者提供了通过蛋白质-DNA复杂结构相互作用的机理见解。但是,这两种类型的数据不容易集成。在这里,我们提出并测试了一种新的建模方法,该方法将PWM与复杂的结构数据结合在一起。在预先调整的蛋白质和DNA粗粒度模型的基础上,我们根据方向相关的电位函数对特定的蛋白质-DNA相互作用PWMcos进行了建模,这使我们能够以前所未有的大规模执行分子动力学模拟。我们表明,PWMcos模型在蛋白质-DNA识别中再现了微妙的特异性。在基因组序列的靶点搜索过程中,TF在高度崎highly的地形上移动,并根据序列偶尔翻动DNA。TATA结合蛋白表现出两种截然不同的结合模式,其中含TATA和无TATA的启动子之间的频率不同。PWMcos是通用的,并且只要其PWM和复杂的结构数据可用,就可以应用于任何蛋白质-DNA相互作用。TATA结合蛋白表现出两种截然不同的结合模式,其中含TATA和无TATA的启动子之间的频率不同。PWMcos是通用的,并且只要其PWM和复杂的结构数据可用,就可以应用于任何蛋白质-DNA相互作用。TATA结合蛋白表现出两种截然不同的结合模式,其中含TATA和无TATA的启动子之间的频率不同。PWMcos是通用的,并且只要其PWM和复杂的结构数据可用,就可以应用于任何蛋白质-DNA相互作用。
更新日期:2018-05-28
中文翻译:

结合测定和结构数据指导的蛋白质-DNA相互作用特异性的动态和结构模型
转录因子(TFs)如何识别其DNA序列通常通过高通量蛋白质结合测定和结构生物学实验进行补充研究。前者可量化TF结合位点对许多DNA序列的特异性,这些序列通常表示为位置权重矩阵(PWM)。后者提供了通过蛋白质-DNA复杂结构相互作用的机理见解。但是,这两种类型的数据不容易集成。在这里,我们提出并测试了一种新的建模方法,该方法将PWM与复杂的结构数据结合在一起。在预先调整的蛋白质和DNA粗粒度模型的基础上,我们根据方向相关的电位函数对特定的蛋白质-DNA相互作用PWMcos进行了建模,这使我们能够以前所未有的大规模执行分子动力学模拟。我们表明,PWMcos模型在蛋白质-DNA识别中再现了微妙的特异性。在基因组序列的靶点搜索过程中,TF在高度崎highly的地形上移动,并根据序列偶尔翻动DNA。TATA结合蛋白表现出两种截然不同的结合模式,其中含TATA和无TATA的启动子之间的频率不同。PWMcos是通用的,并且只要其PWM和复杂的结构数据可用,就可以应用于任何蛋白质-DNA相互作用。TATA结合蛋白表现出两种截然不同的结合模式,其中含TATA和无TATA的启动子之间的频率不同。PWMcos是通用的,并且只要其PWM和复杂的结构数据可用,就可以应用于任何蛋白质-DNA相互作用。TATA结合蛋白表现出两种截然不同的结合模式,其中含TATA和无TATA的启动子之间的频率不同。PWMcos是通用的,并且只要其PWM和复杂的结构数据可用,就可以应用于任何蛋白质-DNA相互作用。










































 京公网安备 11010802027423号
京公网安备 11010802027423号