当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Tuning Gene Activity by Inducible and Targeted Regulation of Gene Expression in Minimal Bacterial Cells
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-05-22 00:00:00 , DOI: 10.1021/acssynbio.8b00028 Ana M Mariscal 1 , Shigeyuki Kakizawa 2, 3 , Jonathan Y Hsu 2, 4 , Kazuki Tanaka 2, 5, 6 , Luis González-González 1 , Alicia Broto 7, 8 , Enrique Querol 1 , Maria Lluch-Senar 7, 8 , Carlos Piñero-Lambea 7, 8 , Lijie Sun 2 , Philip D Weyman 2 , Kim S Wise 2 , Chuck Merryman 2 , Gavin Tse 2, 4 , Adam J Moore 2, 4 , Clyde A Hutchison 2 , Hamilton O Smith 2 , Masaru Tomita 5 , J Craig Venter 2 , John I Glass 2 , Jaume Piñol 1 , Yo Suzuki 2
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-05-22 00:00:00 , DOI: 10.1021/acssynbio.8b00028 Ana M Mariscal 1 , Shigeyuki Kakizawa 2, 3 , Jonathan Y Hsu 2, 4 , Kazuki Tanaka 2, 5, 6 , Luis González-González 1 , Alicia Broto 7, 8 , Enrique Querol 1 , Maria Lluch-Senar 7, 8 , Carlos Piñero-Lambea 7, 8 , Lijie Sun 2 , Philip D Weyman 2 , Kim S Wise 2 , Chuck Merryman 2 , Gavin Tse 2, 4 , Adam J Moore 2, 4 , Clyde A Hutchison 2 , Hamilton O Smith 2 , Masaru Tomita 5 , J Craig Venter 2 , John I Glass 2 , Jaume Piñol 1 , Yo Suzuki 2
Affiliation
|
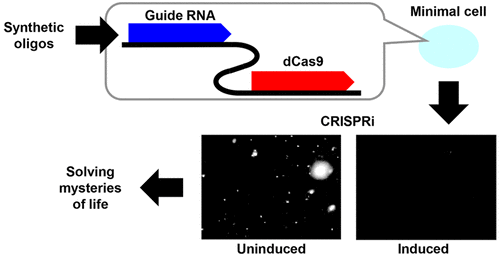
Functional genomics studies in minimal mycoplasma cells enable unobstructed access to some of the most fundamental processes in biology. Conventional transposon bombardment and gene knockout approaches often fail to reveal functions of genes that are essential for viability, where lethality precludes phenotypic characterization. Conditional inactivation of genes is effective for characterizing functions central to cell growth and division, but tools are limited for this purpose in mycoplasmas. Here we demonstrate systems for inducible repression of gene expression based on clustered regularly interspaced short palindromic repeats-mediated interference (CRISPRi) in Mycoplasma pneumoniae and synthetic Mycoplasma mycoides, two organisms with reduced genomes actively used in systems biology studies. In the synthetic cell, we also demonstrate inducible gene expression for the first time. Time-course data suggest rapid kinetics and reversible engagement of CRISPRi. Targeting of six selected endogenous genes with this system results in lowered transcript levels or reduced growth rates that agree with lack or shortage of data in previous transposon bombardment studies, and now produces actual cells to analyze. The ksgA gene encodes a methylase that modifies 16S rRNA, rendering it vulnerable to inhibition by the antibiotic kasugamycin. Targeting the ksgA gene with CRISPRi removes the lethal effect of kasugamycin and enables cell growth, thereby establishing specific and effective gene modulation with our system. The facile methods for conditional gene activation and inactivation in mycoplasmas open the door to systematic dissection of genetic programs at the core of cellular life.
中文翻译:

通过最小细菌细胞中基因表达的诱导性和靶向调节来调节基因活性
最小支原体细胞的功能基因组学研究使人们能够畅通无阻地了解生物学中一些最基本的过程。传统的转座子轰击和基因敲除方法通常无法揭示对于生存至关重要的基因功能,其中致死性妨碍了表型表征。基因的条件失活对于表征细胞生长和分裂的核心功能是有效的,但在支原体中用于此目的的工具有限。在这里,我们展示了基于成簇规则间隔短回文重复介导的干扰(CRISPRi)的基因表达诱导抑制系统,其中肺炎支原体和合成丝状支原体是系统生物学研究中积极使用的两种基因组减少的生物体。在合成细胞中,我们还首次证明了可诱导的基因表达。时程数据表明 CRISPRi 的快速动力学和可逆参与。使用该系统靶向六个选定的内源基因会导致转录物水平降低或生长速率降低,这与之前转座子轰击研究中数据的缺乏或不足相一致,现在可以产生实际的细胞进行分析。 ksgA基因编码一种修饰 16S rRNA 的甲基化酶,使其容易受到抗生素春雷霉素的抑制。用 CRISPRi 靶向ksgA基因可以消除春雷霉素的致死作用并促进细胞生长,从而利用我们的系统建立特异性且有效的基因调节。支原体条件性基因激活和失活的简便方法为系统剖析细胞生命核心的遗传程序打开了大门。
更新日期:2018-05-22
中文翻译:

通过最小细菌细胞中基因表达的诱导性和靶向调节来调节基因活性
最小支原体细胞的功能基因组学研究使人们能够畅通无阻地了解生物学中一些最基本的过程。传统的转座子轰击和基因敲除方法通常无法揭示对于生存至关重要的基因功能,其中致死性妨碍了表型表征。基因的条件失活对于表征细胞生长和分裂的核心功能是有效的,但在支原体中用于此目的的工具有限。在这里,我们展示了基于成簇规则间隔短回文重复介导的干扰(CRISPRi)的基因表达诱导抑制系统,其中肺炎支原体和合成丝状支原体是系统生物学研究中积极使用的两种基因组减少的生物体。在合成细胞中,我们还首次证明了可诱导的基因表达。时程数据表明 CRISPRi 的快速动力学和可逆参与。使用该系统靶向六个选定的内源基因会导致转录物水平降低或生长速率降低,这与之前转座子轰击研究中数据的缺乏或不足相一致,现在可以产生实际的细胞进行分析。 ksgA基因编码一种修饰 16S rRNA 的甲基化酶,使其容易受到抗生素春雷霉素的抑制。用 CRISPRi 靶向ksgA基因可以消除春雷霉素的致死作用并促进细胞生长,从而利用我们的系统建立特异性且有效的基因调节。支原体条件性基因激活和失活的简便方法为系统剖析细胞生命核心的遗传程序打开了大门。









































 京公网安备 11010802027423号
京公网安备 11010802027423号