当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The Catch-22 of Predicting hERG Blockade Using Publicly Accessible Bioactivity Data
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-05-17 00:00:00 , DOI: 10.1021/acs.jcim.8b00150 Vishal B. Siramshetty 1, 2 , Qiaofeng Chen 1, 3 , Prashanth Devarakonda 1 , Robert Preissner 1, 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-05-17 00:00:00 , DOI: 10.1021/acs.jcim.8b00150 Vishal B. Siramshetty 1, 2 , Qiaofeng Chen 1, 3 , Prashanth Devarakonda 1 , Robert Preissner 1, 2
Affiliation
|
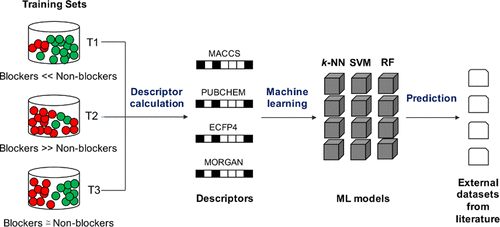
Drug-induced inhibition of the human ether-à-go-go-related gene (hERG)-encoded potassium ion channels can lead to fatal cardiotoxicity. Several marketed drugs and promising drug candidates were recalled because of this concern. Diverse modeling methods ranging from molecular similarity assessment to quantitative structure–activity relationship analysis employing machine learning techniques have been applied to data sets of varying size and composition (number of blockers and nonblockers). In this study, we highlight the challenges involved in the development of a robust classifier for predicting the hERG end point using bioactivity data extracted from the public domain. To this end, three different modeling methods, nearest neighbors, random forests, and support vector machines, were employed to develop predictive models using different molecular descriptors, activity thresholds, and training set compositions. Our models demonstrated superior performance in external validations in comparison with those reported in the previous studies from which the data sets were extracted. The choice of descriptors had little influence on the model performance, with minor exceptions. The criteria used to filter bioactivity data, the activity threshold settings used to separate blockers from nonblockers, and the structural diversity of blockers in training data set were found to be the crucial indicators of model performance. Training sets based on a binary threshold of 1 μM/10 μM to separate blockers (IC50/Ki ≤ 1 μM) from nonblockers (IC50/Ki > 10 μM) provided superior performance in comparison with those defined using a single threshold (1 μM or 10 μM). A major limitation in using the public domain hERG activity data is the abundance of blockers in comparison with nonblockers at usual activity thresholds, since not many studies report the latter.
中文翻译:

使用可公开获得的生物活性数据预测hERG阻断的Catch-22
药物诱导的人源去与相关基因(hERG)编码的钾离子通道的抑制作用可能导致致命的心脏毒性。由于这种担忧,召回了几种市售药物和有前途的候选药物。从分子相似性评估到采用机器学习技术进行定量结构-活性关系分析的各种建模方法,已应用于大小和组成(阻滞剂和非阻滞剂的数量)不同的数据集。在这项研究中,我们重点介绍了使用从公共领域提取的生物活性数据来预测hERG终点的鲁棒分类器的开发所面临的挑战。为此,我们采用了三种不同的建模方法:最近邻居,随机森林和支持向量机,使用不同的分子描述符,活动阈值和训练集组成来开发预测模型。与从中提取数据集的先前研究中报道的模型相比,我们的模型在外部验证中显示出优异的性能。描述符的选择对模型性能几乎没有影响,只有少数例外。发现用于过滤生物活性数据的标准,用于将阻滞剂与非阻滞剂分开的活性阈值设置以及训练数据集中阻滞剂的结构多样性是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 与从中提取数据集的先前研究中报道的模型相比,我们的模型在外部验证中显示出优异的性能。描述符的选择对模型性能几乎没有影响,只有少数例外。发现用于过滤生物活性数据的标准,用于将阻滞剂与非阻滞剂分开的活性阈值设置以及训练数据集中阻滞剂的结构多样性是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 与从中提取数据集的先前研究中报道的模型相比,我们的模型在外部验证中显示出优异的性能。描述符的选择对模型性能几乎没有影响,只有少数例外。发现用于过滤生物活性数据的标准,用于将阻滞剂与非阻滞剂分开的活性阈值设置以及训练数据集中阻滞剂的结构多样性是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 用来区分阻滞剂和非阻滞剂的活动阈值设置,以及训练数据集中阻滞剂的结构多样性被认为是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 用来区分阻滞剂和非阻滞剂的活动阈值设置,以及训练数据集中阻滞剂的结构多样性被认为是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC50 / ķ我≤1μM)从nonblockers(IC 50 / ķ我>10μM)提供与那些使用单个阈值(1μM或10μM)定义的比较优异的性能。使用公共领域hERG活动数据的主要限制是与在常规活动阈值下的非阻滞剂相比,阻滞剂的丰富性,因为没有太多的研究报告后者。
更新日期:2018-05-17
中文翻译:

使用可公开获得的生物活性数据预测hERG阻断的Catch-22
药物诱导的人源去与相关基因(hERG)编码的钾离子通道的抑制作用可能导致致命的心脏毒性。由于这种担忧,召回了几种市售药物和有前途的候选药物。从分子相似性评估到采用机器学习技术进行定量结构-活性关系分析的各种建模方法,已应用于大小和组成(阻滞剂和非阻滞剂的数量)不同的数据集。在这项研究中,我们重点介绍了使用从公共领域提取的生物活性数据来预测hERG终点的鲁棒分类器的开发所面临的挑战。为此,我们采用了三种不同的建模方法:最近邻居,随机森林和支持向量机,使用不同的分子描述符,活动阈值和训练集组成来开发预测模型。与从中提取数据集的先前研究中报道的模型相比,我们的模型在外部验证中显示出优异的性能。描述符的选择对模型性能几乎没有影响,只有少数例外。发现用于过滤生物活性数据的标准,用于将阻滞剂与非阻滞剂分开的活性阈值设置以及训练数据集中阻滞剂的结构多样性是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 与从中提取数据集的先前研究中报道的模型相比,我们的模型在外部验证中显示出优异的性能。描述符的选择对模型性能几乎没有影响,只有少数例外。发现用于过滤生物活性数据的标准,用于将阻滞剂与非阻滞剂分开的活性阈值设置以及训练数据集中阻滞剂的结构多样性是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 与从中提取数据集的先前研究中报道的模型相比,我们的模型在外部验证中显示出优异的性能。描述符的选择对模型性能几乎没有影响,只有少数例外。发现用于过滤生物活性数据的标准,用于将阻滞剂与非阻滞剂分开的活性阈值设置以及训练数据集中阻滞剂的结构多样性是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 用来区分阻滞剂和非阻滞剂的活动阈值设置,以及训练数据集中阻滞剂的结构多样性被认为是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC 用来区分阻滞剂和非阻滞剂的活动阈值设置,以及训练数据集中阻滞剂的结构多样性被认为是模型性能的关键指标。训练集基于1μM/ 10μM的二进制阈值来分离阻断剂(IC50 / ķ我≤1μM)从nonblockers(IC 50 / ķ我>10μM)提供与那些使用单个阈值(1μM或10μM)定义的比较优异的性能。使用公共领域hERG活动数据的主要限制是与在常规活动阈值下的非阻滞剂相比,阻滞剂的丰富性,因为没有太多的研究报告后者。











































 京公网安备 11010802027423号
京公网安备 11010802027423号