当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Toward Spectral Library-Free Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Bacterial Identification.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-05-21 , DOI: 10.1021/acs.jproteome.8b00065 Ding Cheng 1 , Liang Qiao 1 , Peter Horvatovich 2
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-05-21 , DOI: 10.1021/acs.jproteome.8b00065 Ding Cheng 1 , Liang Qiao 1 , Peter Horvatovich 2
Affiliation
|
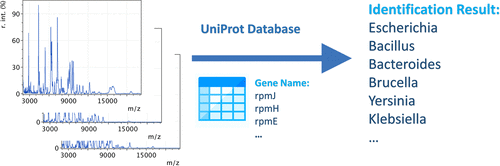
Bacterial identification is of great importance in clinical diagnosis, environmental monitoring, and food safety control. Among various strategies, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has drawn significant interest and has been clinically used. Nevertheless, current bioinformatics solutions use spectral libraries for the identification of bacterial strains. Spectral library generation requires acquisition of MALDI-TOF spectra from monoculture bacterial colonies, which is time-consuming and not possible for many species and strains. We propose a strategy for bacterial typing by MALDI-TOF using protein sequences from public database, that is, UniProt. Ten genes were identified to encode proteins most often observed by MALD-TOF from bacteria through 500 times repeated a 10-fold double cross-validation procedure, using 403 MALDI-TOF spectra corresponding to 14 genera, 81 species, and 403 strains, and the protein sequences of 1276 species in UniProt. The 10 genes were then used to annotate peaks on MALDI-TOF spectra of bacteria for bacterial identification. With the approach, bacteria can be identified at the genus level by searching against a database containing the protein sequences of 42 genera of bacteria from UniProt. Our approach identified 84.1% of the 403 spectra correctly at the genus level. Source code of the algorithm is available at https://github.com/dipcarbon/BacteriaMSLF .
中文翻译:

迈向无谱库的基质辅助激光解吸/电离飞行时间质谱细菌鉴定。
细菌鉴定在临床诊断,环境监测和食品安全控制中非常重要。在各种策略中,基质辅助激光解吸/电离飞行时间质谱(MALDI-TOF MS)引起了人们极大的兴趣,并已在临床上使用。然而,当前的生物信息学解决方案使用光谱库来鉴定细菌菌株。光谱库的生成需要从单培养细菌菌落中获取MALDI-TOF光谱,这很耗时,而且对于许多物种和菌株而言都是不可能的。我们提出了使用来自公共数据库即UniProt的蛋白质序列通过MALDI-TOF进行细菌分型的策略。使用对应于14个属,81个种和403个菌株的403个MALDI-TOF谱图,通过重复10倍双交叉验证程序的500次,鉴定了十个基因来编码MALD-TOF最常从细菌中观察到的蛋白质。 UniProt中1276种的蛋白质序列。然后将这10个基因用于注释细菌的MALDI-TOF谱图上的峰,以进行细菌鉴定。通过这种方法,可以通过检索包含UniProt 42属细菌蛋白质序列的数据库,在属水平上鉴定细菌。我们的方法在属水平上正确地识别了403个光谱中的84.1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。以及UniProt中1276个物种的蛋白质序列。然后将这10个基因用于注释细菌的MALDI-TOF谱图上的峰,以进行细菌鉴定。通过这种方法,可以通过对包含UniProt 42属细菌蛋白质序列的数据库进行搜索来在属水平上鉴定细菌。我们的方法在属水平上正确地识别了403个光谱中的84.1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。以及UniProt中1276个物种的蛋白质序列。然后将这10个基因用于注释细菌的MALDI-TOF谱图上的峰,以进行细菌鉴定。通过这种方法,可以通过对包含UniProt 42属细菌蛋白质序列的数据库进行搜索来在属水平上鉴定细菌。我们的方法在属水平上正确地识别了403个光谱中的84.1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。属属水平的403光谱中正确占1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。属属水平的403光谱中正确占1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。
更新日期:2018-05-22
中文翻译:

迈向无谱库的基质辅助激光解吸/电离飞行时间质谱细菌鉴定。
细菌鉴定在临床诊断,环境监测和食品安全控制中非常重要。在各种策略中,基质辅助激光解吸/电离飞行时间质谱(MALDI-TOF MS)引起了人们极大的兴趣,并已在临床上使用。然而,当前的生物信息学解决方案使用光谱库来鉴定细菌菌株。光谱库的生成需要从单培养细菌菌落中获取MALDI-TOF光谱,这很耗时,而且对于许多物种和菌株而言都是不可能的。我们提出了使用来自公共数据库即UniProt的蛋白质序列通过MALDI-TOF进行细菌分型的策略。使用对应于14个属,81个种和403个菌株的403个MALDI-TOF谱图,通过重复10倍双交叉验证程序的500次,鉴定了十个基因来编码MALD-TOF最常从细菌中观察到的蛋白质。 UniProt中1276种的蛋白质序列。然后将这10个基因用于注释细菌的MALDI-TOF谱图上的峰,以进行细菌鉴定。通过这种方法,可以通过检索包含UniProt 42属细菌蛋白质序列的数据库,在属水平上鉴定细菌。我们的方法在属水平上正确地识别了403个光谱中的84.1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。以及UniProt中1276个物种的蛋白质序列。然后将这10个基因用于注释细菌的MALDI-TOF谱图上的峰,以进行细菌鉴定。通过这种方法,可以通过对包含UniProt 42属细菌蛋白质序列的数据库进行搜索来在属水平上鉴定细菌。我们的方法在属水平上正确地识别了403个光谱中的84.1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。以及UniProt中1276个物种的蛋白质序列。然后将这10个基因用于注释细菌的MALDI-TOF谱图上的峰,以进行细菌鉴定。通过这种方法,可以通过对包含UniProt 42属细菌蛋白质序列的数据库进行搜索来在属水平上鉴定细菌。我们的方法在属水平上正确地识别了403个光谱中的84.1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。属属水平的403光谱中正确占1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。属属水平的403光谱中正确占1%。该算法的源代码可从https://github.com/dipcarbon/BacteriaMSLF获得。









































 京公网安备 11010802027423号
京公网安备 11010802027423号