当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
CypReact: A Software Tool for in Silico Reactant Prediction for Human Cytochrome P450 Enzymes
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-05-08 00:00:00 , DOI: 10.1021/acs.jcim.8b00035 Siyang Tian 1, 2 , Yannick Djoumbou-Feunang 3 , Russell Greiner 1, 2 , David S. Wishart 1, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-05-08 00:00:00 , DOI: 10.1021/acs.jcim.8b00035 Siyang Tian 1, 2 , Yannick Djoumbou-Feunang 3 , Russell Greiner 1, 2 , David S. Wishart 1, 3
Affiliation
|
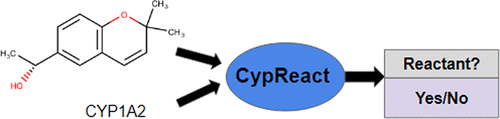
In silico metabolism prediction requires first predicting whether a specific molecule will interact with one or more specific metabolizing enzymes, then predicting the result of each enzymatic reaction. Here, we provide a computational tool, CypReact, for performing this first task of reactant prediction. Specifically, CypReact takes as input an arbitrary molecule (specified as a SMILES string or a standard SDF file) and any one of the nine of the most important human cytochrome P450 (CYP450) enzymes—CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, or CYP3A4—and accurately predicts whether the query molecule will react with that given CYP450 enzyme. Tests of CypReact, conducted over a data set of 1632 molecules (each considered a “plausible” reactant) show that it is very effective, with a (cross-validation) AUROC (area under the receiver operating characteristic curve) of 0.83–0.92. We also show that CypReact performs significantly better than other reactant prediction tools such as ADMET Predictor and (a reactant-predicting extension of) SMARTCyp, whose average AUROCs are 0.75 and 0.53, respectively. We then applied the learned CypReact models to a previously unseen set of molecules and found that our CypReact did even better and still significantly surpassed the performance of SMARTCyp and ADMET Predictor. These results suggest that CypReact could be an important component of a suite of in silico metabolism prediction tools for accurately predicting the products of Phase I, Phase II, and microbial metabolism in humans. CypReact is available at https://bitbucket.org/Leon_Ti/cypreact.
中文翻译:

CypReact:用于人类细胞色素P450酶的硅胶反应物预测的软件工具
在计算机代谢预测中,首先需要预测特定分子是否会与一种或多种特定代谢酶相互作用,然后预测每个酶促反应的结果。在这里,我们提供了一个计算工具CypReact,用于执行反应物预测的第一个任务。具体来说,CypReact将任意分子(指定为SMILES字符串或标准SDF文件)和最重要的人类细胞色素P450(CYP450)九种酶之一(CYP1A2,CYP2A6,CYP2B6,CYP2C8,CYP2C9,CYP2C19)作为输入,CYP2D6,CYP2E1或CYP3A4,并准确预测查询分子是否会与给定的CYP450酶发生反应。对1632个分子(每个被认为是“合理的”反应物)的数据集进行的CypReact测试表明,它非常有效,(交叉验证)AUROC(接收器工作特性曲线下的面积)为0.83-0.92。我们还显示CypReact的性能明显优于其他反应物预测工具,例如ADMET Predictor和SMARTCyp(反应物的预测扩展),后者的平均AUROC分别为0.75和0.53。然后,我们将学习到的CypReact模型应用于以前看不见的一组分子,发现我们的CypReact表现更好,并且仍然大大超过了SMARTCyp和ADMET Predictor的性能。这些结果表明,CypReact可能是一系列计算机代谢预测工具的重要组成部分,可准确预测人的I,II和微生物代谢产物。可以在https://bitbucket.org/Leon_Ti/cypreact上获取CypReact。
更新日期:2018-05-08
中文翻译:

CypReact:用于人类细胞色素P450酶的硅胶反应物预测的软件工具
在计算机代谢预测中,首先需要预测特定分子是否会与一种或多种特定代谢酶相互作用,然后预测每个酶促反应的结果。在这里,我们提供了一个计算工具CypReact,用于执行反应物预测的第一个任务。具体来说,CypReact将任意分子(指定为SMILES字符串或标准SDF文件)和最重要的人类细胞色素P450(CYP450)九种酶之一(CYP1A2,CYP2A6,CYP2B6,CYP2C8,CYP2C9,CYP2C19)作为输入,CYP2D6,CYP2E1或CYP3A4,并准确预测查询分子是否会与给定的CYP450酶发生反应。对1632个分子(每个被认为是“合理的”反应物)的数据集进行的CypReact测试表明,它非常有效,(交叉验证)AUROC(接收器工作特性曲线下的面积)为0.83-0.92。我们还显示CypReact的性能明显优于其他反应物预测工具,例如ADMET Predictor和SMARTCyp(反应物的预测扩展),后者的平均AUROC分别为0.75和0.53。然后,我们将学习到的CypReact模型应用于以前看不见的一组分子,发现我们的CypReact表现更好,并且仍然大大超过了SMARTCyp和ADMET Predictor的性能。这些结果表明,CypReact可能是一系列计算机代谢预测工具的重要组成部分,可准确预测人的I,II和微生物代谢产物。可以在https://bitbucket.org/Leon_Ti/cypreact上获取CypReact。









































 京公网安备 11010802027423号
京公网安备 11010802027423号