当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploration and Comparison of the Geometrical and Physicochemical Properties of an αC Allosteric Pocket in the Structural Kinome
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-04-26 00:00:00 , DOI: 10.1021/acs.jcim.7b00735 Noé Sturm 1 , Annachiara Tinivella 1 , Giulio Rastelli 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-04-26 00:00:00 , DOI: 10.1021/acs.jcim.7b00735 Noé Sturm 1 , Annachiara Tinivella 1 , Giulio Rastelli 1
Affiliation
|
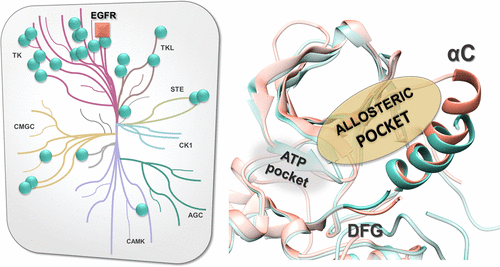
In this work, a comprehensive analysis of the local geometrical and physicochemical properties of a type III allosteric pocket located between the regulatory αC helix and the activation loop of protein kinases was made by comparing available crystal structures in the structural kinome. We first explored the structural kinome to outline the possible conformations of this site. Subsequently we characterized the positions of cocrystallized ligands of the structural kinome with respect to the structural variability of the allosteric site. Then, we searched for kinase structures with similar allosteric site conformation. The search returned 26 kinases with a DFG-in/αC-out conformation potentially prone to bind allosteric inhibitors, as well as different scaffolds that can be useful starting points for the design of new inhibitors. These promising allosteric pockets were probed by performing molecular docking of known active compounds taken from ChEMBL. Interestingly, none of the active compounds reported in ChEMBL had a purely allosteric binding mode, and none of the ATP-competitive ligands had chemical moieties extending into the allosteric pocket in more than two-thirds of the investigated kinases, indicating that the allosteric pocket is accessible but still largely unexplored by available inhibitors. Finally, we compared the physicochemical properties of the allosteric site in the structural kinome and discussed the peculiar and conserved features. These analyses may help the design of allosteric ligands tailored toward the intended kinase(s).
中文翻译:

结构性基因组中αC变构口袋的几何和物理化学性质的探索和比较
在这项工作中,通过比较结构kinome中可用的晶体结构,对位于调节αC螺旋和蛋白激酶激活环之间的III型变构口袋的局部几何和理化性质进行了全面分析。我们首先探索了结构动因,以概述该位点的可能构象。随后,我们针对变构位点的结构变异性表征了结构激酶的共结晶配体的位置。然后,我们搜索了具有相似变构位点构象的激酶结构。搜索返回了26种具有DFG-in /αC-out构象的激酶,这些激酶可能易于结合变构抑制剂,以及不同的支架,这些支架可能是设计新抑制剂的有用起点。通过对取自ChEMBL的已知活性化合物进行分子对接,探索了这些有希望的变构口袋。有趣的是,ChEMBL中报道的所有活性化合物均不具有纯变构结合模式,且ATP竞争性配体中也没有化学成分延伸到超过三分之二的所研究激酶的变构口袋中,这表明变构口袋是可访问的,但仍然很大程度上未被可用的抑制剂开发。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。ChEMBL中报道的所有活性化合物均没有纯变构结合模式,ATP竞争性配体也没有化学成分延伸到超过三分之二的所研究激酶的变构口袋中,这表明该变构口袋可触及,但仍然没有被有效的抑制剂开发。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。ChEMBL中报道的所有活性化合物均没有纯变构结合模式,ATP竞争性配体也没有化学成分延伸到超过三分之二的所研究激酶的变构口袋中,这表明该变构口袋可触及,但仍然没有被有效的抑制剂开发。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。表明变构口袋是可接近的,但在很大程度上仍未被可用的抑制剂研究。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。表明变构口袋是可接近的,但在很大程度上仍未被可用的抑制剂研究。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。
更新日期:2018-04-26
中文翻译:

结构性基因组中αC变构口袋的几何和物理化学性质的探索和比较
在这项工作中,通过比较结构kinome中可用的晶体结构,对位于调节αC螺旋和蛋白激酶激活环之间的III型变构口袋的局部几何和理化性质进行了全面分析。我们首先探索了结构动因,以概述该位点的可能构象。随后,我们针对变构位点的结构变异性表征了结构激酶的共结晶配体的位置。然后,我们搜索了具有相似变构位点构象的激酶结构。搜索返回了26种具有DFG-in /αC-out构象的激酶,这些激酶可能易于结合变构抑制剂,以及不同的支架,这些支架可能是设计新抑制剂的有用起点。通过对取自ChEMBL的已知活性化合物进行分子对接,探索了这些有希望的变构口袋。有趣的是,ChEMBL中报道的所有活性化合物均不具有纯变构结合模式,且ATP竞争性配体中也没有化学成分延伸到超过三分之二的所研究激酶的变构口袋中,这表明变构口袋是可访问的,但仍然很大程度上未被可用的抑制剂开发。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。ChEMBL中报道的所有活性化合物均没有纯变构结合模式,ATP竞争性配体也没有化学成分延伸到超过三分之二的所研究激酶的变构口袋中,这表明该变构口袋可触及,但仍然没有被有效的抑制剂开发。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。ChEMBL中报道的所有活性化合物均没有纯变构结合模式,ATP竞争性配体也没有化学成分延伸到超过三分之二的所研究激酶的变构口袋中,这表明该变构口袋可触及,但仍然没有被有效的抑制剂开发。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。表明变构口袋是可接近的,但在很大程度上仍未被可用的抑制剂研究。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。表明变构口袋是可接近的,但在很大程度上仍未被可用的抑制剂研究。最后,我们比较了结构性基因组中变构位点的理化性质,并讨论了其独特和保守的特征。这些分析可以帮助设计针对预期激酶的变构配体。











































 京公网安备 11010802027423号
京公网安备 11010802027423号