Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structure and regulation of the human INO80–nucleosome complex
Nature ( IF 50.5 ) Pub Date : 2018-04-01 , DOI: 10.1038/s41586-018-0021-6 Rafael Ayala 1 , Oliver Willhoft 1 , Ricardo J Aramayo 1 , Martin Wilkinson 1 , Elizabeth A McCormack 1 , Lorraine Ocloo 1 , Dale B Wigley 1 , Xiaodong Zhang 1
Nature ( IF 50.5 ) Pub Date : 2018-04-01 , DOI: 10.1038/s41586-018-0021-6 Rafael Ayala 1 , Oliver Willhoft 1 , Ricardo J Aramayo 1 , Martin Wilkinson 1 , Elizabeth A McCormack 1 , Lorraine Ocloo 1 , Dale B Wigley 1 , Xiaodong Zhang 1
Affiliation
|
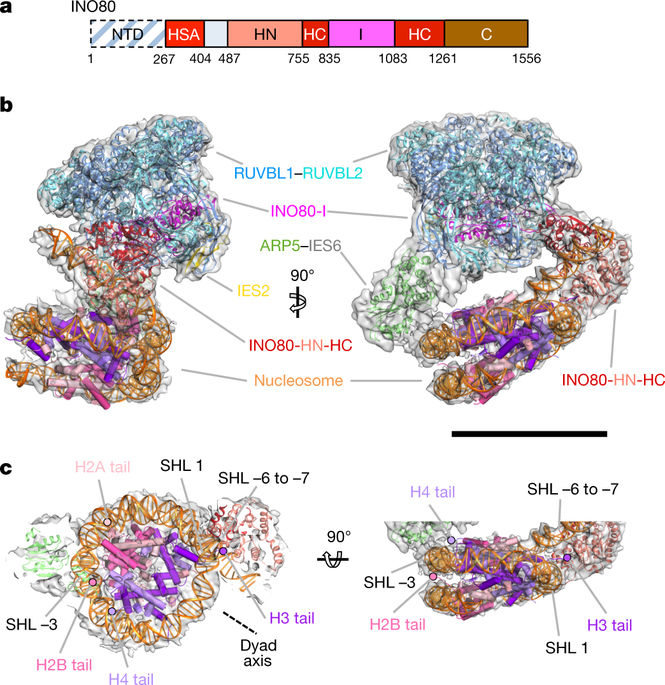
Access to DNA within nucleosomes is required for a variety of processes in cells including transcription, replication and repair. Consequently, cells encode multiple systems that remodel nucleosomes. These complexes can be simple, involving one or a few protein subunits, or more complicated multi-subunit machines1. Biochemical studies2–4 have placed the motor domains of several chromatin remodellers in the superhelical location 2 region of the nucleosome. Structural studies of yeast Chd1 and Snf2—a subunit in the complex with the capacity to remodel the structure of chromatin (RSC)—in complex with nucleosomes5–7 have provided insights into the basic mechanism of nucleosome sliding performed by these complexes. However, how larger, multi-subunit remodelling complexes such as INO80 interact with nucleosomes and how remodellers carry out functions such as nucleosome sliding8, histone exchange9 and nucleosome spacing10–12 remain poorly understood. Although some remodellers work as monomers13, others work as highly cooperative dimers11, 14, 15. Here we present the structure of the human INO80 chromatin remodeller with a bound nucleosome, which reveals that INO80 interacts with nucleosomes in a previously undescribed manner: the motor domains are located on the DNA at the entry point to the nucleosome, rather than at superhelical location 2. The ARP5–IES6 module of INO80 makes additional contacts on the opposite side of the nucleosome. This arrangement enables the histone H3 tails of the nucleosome to have a role in the regulation of the activities of the INO80 motor domain—unlike in other characterized remodellers, for which H4 tails have been shown to regulate the motor domains.Cryo-electron microscopy structure of the human INO80 chromatin remodeller in complex with a bound nucleosome reveals that its motor domains are located at the DNA wrap around the histone core.
中文翻译:

人类 INO80-核小体复合物的结构和调控
细胞内的多种过程(包括转录、复制和修复)都需要接触核小体内的 DNA。因此,细胞编码多个重塑核小体的系统。这些复合物可以很简单,涉及一个或几个蛋白质亚基,也可以是更复杂的多亚基机器1。生化研究 2-4 将几种染色质重塑因子的运动域置于核小体的超螺旋位置 2 区域。对酵母 Chd1 和 Snf2(复合物中的一个亚基,具有重塑染色质 (RSC) 结构的能力)与核小体 5-7 复合物的结构研究,为了解这些复合物进行核小体滑动的基本机制提供了见解。然而,诸如 INO80 等较大的多亚基重塑复合物如何与核小体相互作用,以及重塑者如何执行核小体滑动 8、组蛋白交换 9 和核小体间距 10-12 等功能,仍然知之甚少。虽然一些重塑剂以单体的形式发挥作用13,但其他重塑剂以高度合作的二聚体的形式发挥作用11、14、15。在这里,我们展示了具有结合核小体的人类 INO80 染色质重塑剂的结构,这揭示了 INO80 以以前未描述的方式与核小体相互作用:运动域位于 DNA 上核小体的入口点,而不是超螺旋位置 2。INO80 的 ARP5-IES6 模块在核小体的另一侧产生额外的接触。这种排列使得核小体的组蛋白 H3 尾能够在调节 INO80 运动域的活动中发挥作用,这与其他特征重塑者不同,后者的 H4 尾已被证明可以调节运动域。 冷冻电子显微镜结构与结合核小体复合的人类 INO80 染色质重塑蛋白的研究表明,其运动结构域位于组蛋白核心周围的 DNA 包裹处。
更新日期:2018-04-01
中文翻译:

人类 INO80-核小体复合物的结构和调控
细胞内的多种过程(包括转录、复制和修复)都需要接触核小体内的 DNA。因此,细胞编码多个重塑核小体的系统。这些复合物可以很简单,涉及一个或几个蛋白质亚基,也可以是更复杂的多亚基机器1。生化研究 2-4 将几种染色质重塑因子的运动域置于核小体的超螺旋位置 2 区域。对酵母 Chd1 和 Snf2(复合物中的一个亚基,具有重塑染色质 (RSC) 结构的能力)与核小体 5-7 复合物的结构研究,为了解这些复合物进行核小体滑动的基本机制提供了见解。然而,诸如 INO80 等较大的多亚基重塑复合物如何与核小体相互作用,以及重塑者如何执行核小体滑动 8、组蛋白交换 9 和核小体间距 10-12 等功能,仍然知之甚少。虽然一些重塑剂以单体的形式发挥作用13,但其他重塑剂以高度合作的二聚体的形式发挥作用11、14、15。在这里,我们展示了具有结合核小体的人类 INO80 染色质重塑剂的结构,这揭示了 INO80 以以前未描述的方式与核小体相互作用:运动域位于 DNA 上核小体的入口点,而不是超螺旋位置 2。INO80 的 ARP5-IES6 模块在核小体的另一侧产生额外的接触。这种排列使得核小体的组蛋白 H3 尾能够在调节 INO80 运动域的活动中发挥作用,这与其他特征重塑者不同,后者的 H4 尾已被证明可以调节运动域。 冷冻电子显微镜结构与结合核小体复合的人类 INO80 染色质重塑蛋白的研究表明,其运动结构域位于组蛋白核心周围的 DNA 包裹处。










































 京公网安备 11010802027423号
京公网安备 11010802027423号