当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dynamics, Conformational Entropy, and Frustration in Protein–Protein Interactions Involving an Intrinsically Disordered Protein Domain
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2018-04-03 00:00:00 , DOI: 10.1021/acschembio.7b01105 Ida Lindström 1 , Jakob Dogan 1
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2018-04-03 00:00:00 , DOI: 10.1021/acschembio.7b01105 Ida Lindström 1 , Jakob Dogan 1
Affiliation
|
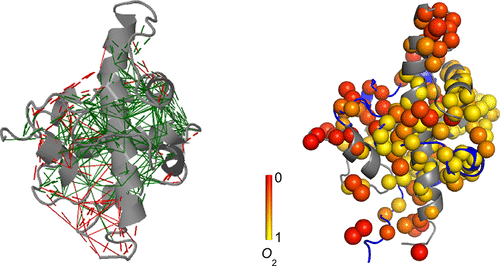
Intrinsically disordered proteins (IDPs) are abundant in the eukaryotic proteome. However, little is known about the role of subnanosecond dynamics and the conformational entropy that it represents in protein–protein interactions involving IDPs. Using nuclear magnetic resonance side chain and backbone relaxation, stopped-flow kinetics, isothermal titration calorimetry, and computational studies, we have characterized the interaction between the globular TAZ1 domain of the CREB binding protein and the intrinsically disordered transactivation domain of STAT2 (TAD-STAT2). We show that the TAZ1/TAD-STAT2 complex retains considerable subnanosecond motions, with TAD-STAT2 undergoing only a partial disorder-to-order transition. We report here the first experimental determination of the conformational entropy change for both binding partners in an IDP binding interaction and find that the total change even exceeds in magnitude the binding enthalpy and is comparable to the contribution from the hydrophobic effect, demonstrating its importance in the binding energetics. Furthermore, we show that the conformational entropy change for TAZ1 is also instrumental in maintaining a biologically meaningful binding affinity. Strikingly, a spatial clustering of very high amplitude motions and a cluster of more rigid sites in the complex exist, which through computational studies we found to overlap with regions that experience energetic frustration and are less frustrated, respectively. Thus, the residual dynamics in the bound state could be necessary for faster dissociation, which is important for proteins that interact with multiple binding partners.
中文翻译:

动力学,构象熵和挫折中涉及本质上混乱的蛋白质域的蛋白质-蛋白质相互作用。
内在无序的蛋白质(IDPs)在真核蛋白质组中是丰富的。然而,人们对亚纳秒动力学的作用及其在涉及IDP的蛋白质相互作用中所表现出的构象熵知之甚少。利用核磁共振侧链和主链弛豫,停流动力学,等温滴定量热法和计算研究,我们表征了CREB结合蛋白的球状TAZ1结构域与STAT2(TAD-STAT2)的固有无序反式激活结构域之间的相互作用。 )。我们显示,TAZ1 / TAD-STAT2复合物保留了相当的亚纳秒运动,而TAD-STAT2仅经历了部分无序到有序的过渡。我们在这里报告了IDP结合相互作用中两个结合配偶体的构象熵变化的首次实验测定,发现总变化量甚至超过了结合焓,可与疏水作用相媲美,证明了其在疏水作用中的重要性。约束能量学。此外,我们表明,TAZ1的构象熵变化在维持生物学上有意义的结合亲和力中也起着重要作用。引人注目的是,在复合物中存在着一个非常高幅度的运动的空间聚类和一个较刚性的位置的聚类,通过计算研究,我们发现它们分别与遭受精力充沛的挫败和较少挫败的区域重叠。因此,在束缚状态下的残留动力学可能对于更快的离解是必要的,
更新日期:2018-04-03
中文翻译:

动力学,构象熵和挫折中涉及本质上混乱的蛋白质域的蛋白质-蛋白质相互作用。
内在无序的蛋白质(IDPs)在真核蛋白质组中是丰富的。然而,人们对亚纳秒动力学的作用及其在涉及IDP的蛋白质相互作用中所表现出的构象熵知之甚少。利用核磁共振侧链和主链弛豫,停流动力学,等温滴定量热法和计算研究,我们表征了CREB结合蛋白的球状TAZ1结构域与STAT2(TAD-STAT2)的固有无序反式激活结构域之间的相互作用。 )。我们显示,TAZ1 / TAD-STAT2复合物保留了相当的亚纳秒运动,而TAD-STAT2仅经历了部分无序到有序的过渡。我们在这里报告了IDP结合相互作用中两个结合配偶体的构象熵变化的首次实验测定,发现总变化量甚至超过了结合焓,可与疏水作用相媲美,证明了其在疏水作用中的重要性。约束能量学。此外,我们表明,TAZ1的构象熵变化在维持生物学上有意义的结合亲和力中也起着重要作用。引人注目的是,在复合物中存在着一个非常高幅度的运动的空间聚类和一个较刚性的位置的聚类,通过计算研究,我们发现它们分别与遭受精力充沛的挫败和较少挫败的区域重叠。因此,在束缚状态下的残留动力学可能对于更快的离解是必要的,











































 京公网安备 11010802027423号
京公网安备 11010802027423号