Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis
Nature ( IF 50.5 ) Pub Date : 2018-03-28 , DOI: 10.1038/nature25985 Megan Sjodt , Kelly Brock , Genevieve Dobihal , Patricia D. A. Rohs , Anna G. Green , Thomas A. Hopf , Alexander J. Meeske , Veerasak Srisuknimit , Daniel Kahne , Suzanne Walker , Debora S. Marks , Thomas G. Bernhardt , David Z. Rudner , Andrew C. Kruse
Nature ( IF 50.5 ) Pub Date : 2018-03-28 , DOI: 10.1038/nature25985 Megan Sjodt , Kelly Brock , Genevieve Dobihal , Patricia D. A. Rohs , Anna G. Green , Thomas A. Hopf , Alexander J. Meeske , Veerasak Srisuknimit , Daniel Kahne , Suzanne Walker , Debora S. Marks , Thomas G. Bernhardt , David Z. Rudner , Andrew C. Kruse
|
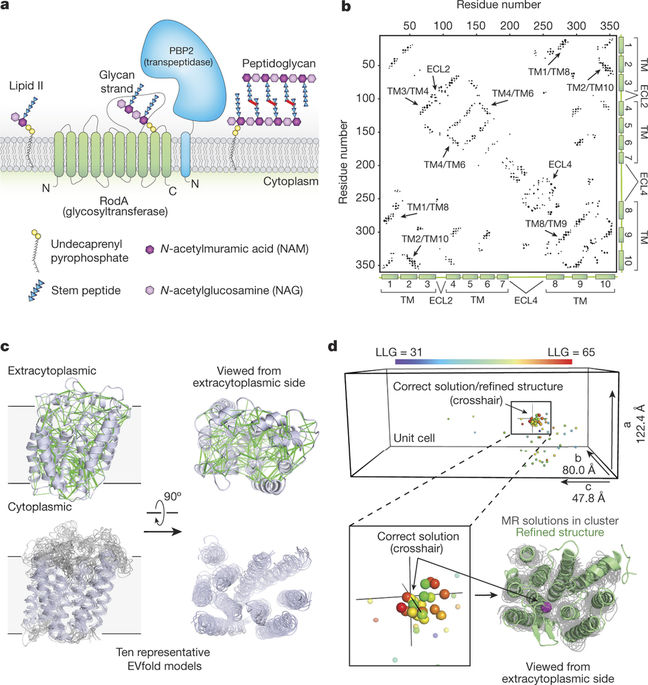
The shape, elongation, division and sporulation (SEDS) proteins are a large family of ubiquitous and essential transmembrane enzymes with critical roles in bacterial cell wall biology. The exact function of SEDS proteins was for a long time poorly understood, but recent work has revealed that the prototypical SEDS family member RodA is a peptidoglycan polymerase—a role previously attributed exclusively to members of the penicillin-binding protein family. This discovery has made RodA and other SEDS proteins promising targets for the development of next-generation antibiotics. However, little is known regarding the molecular basis of SEDS activity, and no structural data are available for RodA or any homologue thereof. Here we report the crystal structure of Thermus thermophilus RodA at a resolution of 2.9 Å, determined using evolutionary covariance-based fold prediction to enable molecular replacement. The structure reveals a ten-pass transmembrane fold with large extracellular loops, one of which is partially disordered. The protein contains a highly conserved cavity in the transmembrane domain, reminiscent of ligand-binding sites in transmembrane receptors. Mutagenesis experiments in Bacillus subtilis and Escherichia coli show that perturbation of this cavity abolishes RodA function both in vitro and in vivo, indicating that this cavity is catalytically essential. These results provide a framework for understanding bacterial cell wall synthesis and SEDS protein function.
中文翻译:

通过进化耦合分析解析肽聚糖聚合酶 RodA 的结构
形状、伸长、分裂和孢子形成 (SEDS) 蛋白是一大类普遍存在且必不可少的跨膜酶,在细菌细胞壁生物学中具有关键作用。长期以来,人们对 SEDS 蛋白的确切功能知之甚少,但最近的研究表明,典型的 SEDS 家族成员 RodA 是一种肽聚糖聚合酶——以前这种作用仅归因于青霉素结合蛋白家族的成员。这一发现使 RodA 和其他 SEDS 蛋白成为开发下一代抗生素的有希望的目标。然而,关于 SEDS 活性的分子基础知之甚少,也没有 RodA 或其任何同系物的结构数据。在这里,我们以 2.9 Å 的分辨率报告了嗜热菌 RodA 的晶体结构,使用基于进化协方差的折叠预测来确定,以实现分子替换。该结构显示出具有大细胞外环的十次跨膜折叠,其中一个是部分无序的。该蛋白质在跨膜结构域中包含一个高度保守的空腔,让人联想到跨膜受体中的配体结合位点。枯草芽孢杆菌和大肠杆菌中的诱变实验表明,该腔的扰动在体外和体内都消除了 RodA 功能,表明该腔具有催化作用。这些结果为理解细菌细胞壁合成和 SEDS 蛋白功能提供了一个框架。该蛋白质在跨膜结构域中包含一个高度保守的空腔,让人联想到跨膜受体中的配体结合位点。枯草芽孢杆菌和大肠杆菌中的诱变实验表明,该腔的扰动在体外和体内都消除了 RodA 功能,表明该腔具有催化作用。这些结果为理解细菌细胞壁合成和 SEDS 蛋白功能提供了一个框架。该蛋白质在跨膜结构域中包含一个高度保守的空腔,让人联想到跨膜受体中的配体结合位点。枯草芽孢杆菌和大肠杆菌中的诱变实验表明,该腔的扰动在体外和体内都消除了 RodA 功能,表明该腔具有催化作用。这些结果为理解细菌细胞壁合成和 SEDS 蛋白功能提供了一个框架。
更新日期:2018-03-28
中文翻译:

通过进化耦合分析解析肽聚糖聚合酶 RodA 的结构
形状、伸长、分裂和孢子形成 (SEDS) 蛋白是一大类普遍存在且必不可少的跨膜酶,在细菌细胞壁生物学中具有关键作用。长期以来,人们对 SEDS 蛋白的确切功能知之甚少,但最近的研究表明,典型的 SEDS 家族成员 RodA 是一种肽聚糖聚合酶——以前这种作用仅归因于青霉素结合蛋白家族的成员。这一发现使 RodA 和其他 SEDS 蛋白成为开发下一代抗生素的有希望的目标。然而,关于 SEDS 活性的分子基础知之甚少,也没有 RodA 或其任何同系物的结构数据。在这里,我们以 2.9 Å 的分辨率报告了嗜热菌 RodA 的晶体结构,使用基于进化协方差的折叠预测来确定,以实现分子替换。该结构显示出具有大细胞外环的十次跨膜折叠,其中一个是部分无序的。该蛋白质在跨膜结构域中包含一个高度保守的空腔,让人联想到跨膜受体中的配体结合位点。枯草芽孢杆菌和大肠杆菌中的诱变实验表明,该腔的扰动在体外和体内都消除了 RodA 功能,表明该腔具有催化作用。这些结果为理解细菌细胞壁合成和 SEDS 蛋白功能提供了一个框架。该蛋白质在跨膜结构域中包含一个高度保守的空腔,让人联想到跨膜受体中的配体结合位点。枯草芽孢杆菌和大肠杆菌中的诱变实验表明,该腔的扰动在体外和体内都消除了 RodA 功能,表明该腔具有催化作用。这些结果为理解细菌细胞壁合成和 SEDS 蛋白功能提供了一个框架。该蛋白质在跨膜结构域中包含一个高度保守的空腔,让人联想到跨膜受体中的配体结合位点。枯草芽孢杆菌和大肠杆菌中的诱变实验表明,该腔的扰动在体外和体内都消除了 RodA 功能,表明该腔具有催化作用。这些结果为理解细菌细胞壁合成和 SEDS 蛋白功能提供了一个框架。











































 京公网安备 11010802027423号
京公网安备 11010802027423号