Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Whole-organism clone tracing using single-cell sequencing
Nature ( IF 50.5 ) Pub Date : 2018-03-28 , DOI: 10.1038/nature25969 Anna Alemany , Maria Florescu , Chloé S. Baron , Josi Peterson-Maduro , Alexander van Oudenaarden
Nature ( IF 50.5 ) Pub Date : 2018-03-28 , DOI: 10.1038/nature25969 Anna Alemany , Maria Florescu , Chloé S. Baron , Josi Peterson-Maduro , Alexander van Oudenaarden
|
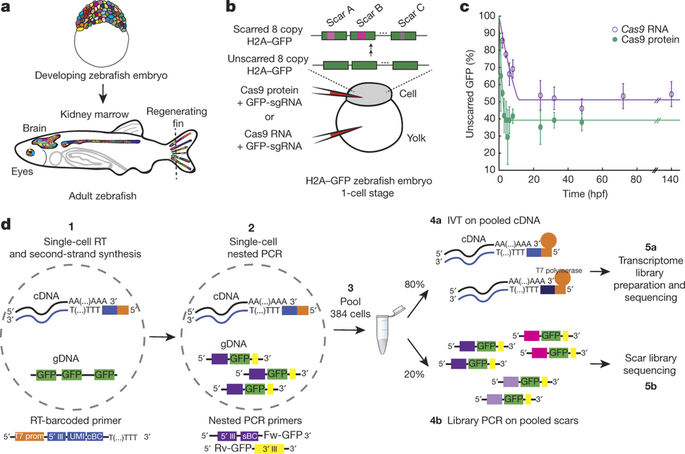
Embryonic development is a crucial period in the life of a multicellular organism, during which limited sets of embryonic progenitors produce all cells in the adult body. Determining which fate these progenitors acquire in adult tissues requires the simultaneous measurement of clonal history and cell identity at single-cell resolution, which has been a major challenge. Clonal history has traditionally been investigated by microscopically tracking cells during development, monitoring the heritable expression of genetically encoded fluorescent proteins and, more recently, using next-generation sequencing technologies that exploit somatic mutations, microsatellite instability, transposon tagging, viral barcoding, CRISPR–Cas9 genome editing and Cre–loxP recombination. Single-cell transcriptomics provides a powerful platform for unbiased cell-type classification. Here we present ScarTrace, a single-cell sequencing strategy that enables the simultaneous quantification of clonal history and cell type for thousands of cells obtained from different organs of the adult zebrafish. Using ScarTrace, we show that a small set of multipotent embryonic progenitors generate all haematopoietic cells in the kidney marrow, and that many progenitors produce specific cell types in the eyes and brain. In addition, we study when embryonic progenitors commit to the left or right eye. ScarTrace reveals that epidermal and mesenchymal cells in the caudal fin arise from the same progenitors, and that osteoblast-restricted precursors can produce mesenchymal cells during regeneration. Furthermore, we identify resident immune cells in the fin with a distinct clonal origin from other blood cell types. We envision that similar approaches will have major applications in other experimental systems, in which the matching of embryonic clonal origin to adult cell type will ultimately allow reconstruction of how the adult body is built from a single cell.
中文翻译:

使用单细胞测序进行全生物克隆追踪
胚胎发育是多细胞生物生命中的关键时期,在此期间,有限的胚胎祖细胞会产生成人体内的所有细胞。确定这些祖细胞在成人组织中获得的命运需要以单细胞分辨率同时测量克隆历史和细胞身份,这是一个重大挑战。克隆历史传统上通过在发育过程中显微镜跟踪细胞、监测遗传编码荧光蛋白的遗传表达以及最近使用利用体细胞突变、微卫星不稳定性、转座子标记、病毒条形码、CRISPR-Cas9 的下一代测序技术来研究基因组编辑和 Cre-loxP 重组。单细胞转录组学为无偏见的细胞类型分类提供了一个强大的平台。在这里,我们介绍了 ScarTrace,这是一种单细胞测序策略,可以同时量化从成年斑马鱼不同器官获得的数千个细胞的克隆历史和细胞类型。使用 ScarTrace,我们展示了一小组多能胚胎祖细胞在肾骨髓中生成所有造血细胞,并且许多祖细胞在眼睛和大脑中生成特定类型的细胞。此外,我们研究胚胎祖细胞何时出现在左眼或右眼。ScarTrace 揭示尾鳍中的表皮和间充质细胞来自相同的祖细胞,并且成骨细胞限制的前体可以在再生过程中产生间充质细胞。此外,我们确定了鳍中的常驻免疫细胞具有与其他血细胞类型不同的克隆来源。我们设想类似的方法将在其他实验系统中具有重要应用,其中胚胎克隆起源与成体细胞类型的匹配将最终允许重建成体是如何从单个细胞构建的。
更新日期:2018-03-28
中文翻译:

使用单细胞测序进行全生物克隆追踪
胚胎发育是多细胞生物生命中的关键时期,在此期间,有限的胚胎祖细胞会产生成人体内的所有细胞。确定这些祖细胞在成人组织中获得的命运需要以单细胞分辨率同时测量克隆历史和细胞身份,这是一个重大挑战。克隆历史传统上通过在发育过程中显微镜跟踪细胞、监测遗传编码荧光蛋白的遗传表达以及最近使用利用体细胞突变、微卫星不稳定性、转座子标记、病毒条形码、CRISPR-Cas9 的下一代测序技术来研究基因组编辑和 Cre-loxP 重组。单细胞转录组学为无偏见的细胞类型分类提供了一个强大的平台。在这里,我们介绍了 ScarTrace,这是一种单细胞测序策略,可以同时量化从成年斑马鱼不同器官获得的数千个细胞的克隆历史和细胞类型。使用 ScarTrace,我们展示了一小组多能胚胎祖细胞在肾骨髓中生成所有造血细胞,并且许多祖细胞在眼睛和大脑中生成特定类型的细胞。此外,我们研究胚胎祖细胞何时出现在左眼或右眼。ScarTrace 揭示尾鳍中的表皮和间充质细胞来自相同的祖细胞,并且成骨细胞限制的前体可以在再生过程中产生间充质细胞。此外,我们确定了鳍中的常驻免疫细胞具有与其他血细胞类型不同的克隆来源。我们设想类似的方法将在其他实验系统中具有重要应用,其中胚胎克隆起源与成体细胞类型的匹配将最终允许重建成体是如何从单个细胞构建的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号