当前位置:
X-MOL 学术
›
ACS Chem. Neurosci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Proteomic Characterization of the Neural Ectoderm Fated Cell Clones in the Xenopus laevis Embryo by High-Resolution Mass Spectrometry.
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2018-04-05 , DOI: 10.1021/acschemneuro.7b00525 Aparna B Baxi 1, 2 , Camille Lombard-Banek 1 , Sally A Moody 2 , Peter Nemes 1, 2
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2018-04-05 , DOI: 10.1021/acschemneuro.7b00525 Aparna B Baxi 1, 2 , Camille Lombard-Banek 1 , Sally A Moody 2 , Peter Nemes 1, 2
Affiliation
|
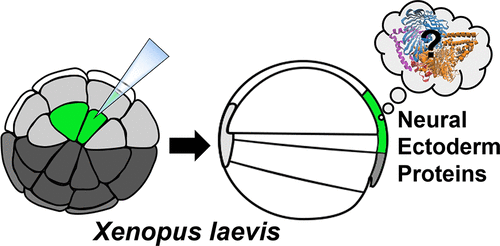
The molecular program by which embryonic ectoderm is induced to form neural tissue is essential to understanding normal and impaired development of the central nervous system. Xenopus has been a powerful vertebrate model in which to elucidate this process. However, abundant vitellogenin (yolk) proteins in cells of the early Xenopus embryo interfere with protein detection by high-resolution mass spectrometry (HRMS), the technology of choice for identifying these gene products. Here, we systematically evaluated strategies of bottom-up proteomics to enhance proteomic detection from the neural ectoderm (NE) of X. laevis using nanoflow high-performance liquid chromatography (nanoLC) HRMS. From whole embryos, high-pH fractionation prior to nanoLC-HRMS yielded 1319 protein groups vs 762 proteins without fractionation (control). Compared to 702 proteins from dorsal halves of embryos (control), 1881 proteins were identified after yolk platelets were depleted via sucrose-gradient centrifugation. We combined these approaches to characterize protein expression in the NE of the early embryo. To guide microdissection of the NE tissues from the gastrula (stage 10), their precursor (midline dorsal-animal, or D111) cells were fate-mapped from the 32-cell embryo using a fluorescent lineage tracer. HRMS of the cell clones identified 2363 proteins, including 147 phosphoproteins (without phosphoprotein enrichment), transcription factors, and members from pathways of cellular signaling. In reference to transcriptomic maps of the developing X. laevis, 76 proteins involved in signaling pathways were gene matched to transcripts with known enrichment in the neural plate. Besides a protocol, this work provides qualitative proteomic data on the early developing NE.
中文翻译:

爪蟾胚胎胚胎中神经外胚层细胞克隆的蛋白质组学表征。
诱导胚胎外胚层形成神经组织的分子程序对于理解中枢神经系统的正常和受损发育至关重要。爪蟾已经成为阐明这一过程的强有力的脊椎动物模型。但是,非洲爪蟾早期胚胎细胞中大量的卵黄蛋白原(卵黄)蛋白干扰了高分辨率质谱(HRMS)的蛋白质检测,高分辨率质谱是鉴定这些基因产物的首选技术。在这里,我们系统地评估了自底向上蛋白质组学的策略,以使用纳流高效液相色谱(nanoLC)HRMS从X. laevis的神经外胚层(NE)增强蛋白质组学检测。从整个胚胎中,在nanoLC-HRMS之前进行高pH分离可得到1319个蛋白质组,而对不分离的762个蛋白进行了分离(对照)。与来自胚胎后半部分的702种蛋白质(对照)相比,通过蔗糖梯度离心去除蛋黄血小板后,鉴定出了1881种蛋白质。我们结合这些方法来表征蛋白质在早期胚胎的NE中的表达。为了指导胃小肠NE组织的显微解剖(第10期),使用荧光谱系示踪剂从32细胞胚胎中确定了它们的前体细胞(中线背侧动物或D111)。细胞克隆的HRMS鉴定出2363种蛋白质,包括147种磷蛋白(无磷蛋白富集),转录因子和来自细胞信号通路的成员。参照发育中的X. laevis的转录组图谱,将与信号通路有关的76种蛋白质与在神经板中已知富集的转录本进行基因匹配。
更新日期:2018-03-26
中文翻译:

爪蟾胚胎胚胎中神经外胚层细胞克隆的蛋白质组学表征。
诱导胚胎外胚层形成神经组织的分子程序对于理解中枢神经系统的正常和受损发育至关重要。爪蟾已经成为阐明这一过程的强有力的脊椎动物模型。但是,非洲爪蟾早期胚胎细胞中大量的卵黄蛋白原(卵黄)蛋白干扰了高分辨率质谱(HRMS)的蛋白质检测,高分辨率质谱是鉴定这些基因产物的首选技术。在这里,我们系统地评估了自底向上蛋白质组学的策略,以使用纳流高效液相色谱(nanoLC)HRMS从X. laevis的神经外胚层(NE)增强蛋白质组学检测。从整个胚胎中,在nanoLC-HRMS之前进行高pH分离可得到1319个蛋白质组,而对不分离的762个蛋白进行了分离(对照)。与来自胚胎后半部分的702种蛋白质(对照)相比,通过蔗糖梯度离心去除蛋黄血小板后,鉴定出了1881种蛋白质。我们结合这些方法来表征蛋白质在早期胚胎的NE中的表达。为了指导胃小肠NE组织的显微解剖(第10期),使用荧光谱系示踪剂从32细胞胚胎中确定了它们的前体细胞(中线背侧动物或D111)。细胞克隆的HRMS鉴定出2363种蛋白质,包括147种磷蛋白(无磷蛋白富集),转录因子和来自细胞信号通路的成员。参照发育中的X. laevis的转录组图谱,将与信号通路有关的76种蛋白质与在神经板中已知富集的转录本进行基因匹配。











































 京公网安备 11010802027423号
京公网安备 11010802027423号