当前位置:
X-MOL 学术
›
ACS Infect. Dis.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Antibiotic Combinations That Enable One-Step, Targeted Mutagenesis of Chromosomal Genes
ACS Infectious Diseases ( IF 4.0 ) Pub Date : 2018-03-13 00:00:00 , DOI: 10.1021/acsinfecdis.8b00017 Wonsik Lee 1 , Truc Do 1 , Ge Zhang 1, 2 , Daniel Kahne 2 , Timothy C. Meredith 1 , Suzanne Walker 1
ACS Infectious Diseases ( IF 4.0 ) Pub Date : 2018-03-13 00:00:00 , DOI: 10.1021/acsinfecdis.8b00017 Wonsik Lee 1 , Truc Do 1 , Ge Zhang 1, 2 , Daniel Kahne 2 , Timothy C. Meredith 1 , Suzanne Walker 1
Affiliation
|
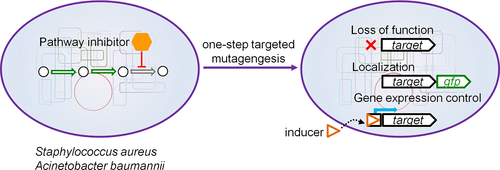
Targeted modification of bacterial chromosomes is necessary to understand new drug targets, investigate virulence factors, elucidate cell physiology, and validate results of -omics-based approaches. For some bacteria, reverse genetics remains a major bottleneck to progress in research. Here, we describe a compound-centric strategy that combines new negative selection markers with known positive selection markers to achieve simple, efficient one-step genome engineering of bacterial chromosomes. The method was inspired by the observation that certain nonessential metabolic pathways contain essential late steps, suggesting that antibiotics targeting a late step can be used to select for the absence of genes that control flux into the pathway. Guided by this hypothesis, we have identified antibiotic/counterselectable markers to accelerate reverse engineering of two increasingly antibiotic-resistant pathogens, Staphylococcus aureus and Acinetobacter baumannii. For S. aureus, we used wall teichoic acid biosynthesis inhibitors to select for the absence of tarO and for A. baumannii, we used colistin to select for the absence of lpxC. We have obtained desired gene deletions, gene fusions, and promoter swaps in a single plating step with perfect efficiency. Our method can also be adapted to generate markerless deletions of genes using FLP recombinase. The tools described here will accelerate research on two important pathogens, and the concept we outline can be readily adapted to any organism for which a suitable target pathway can be identified.
中文翻译:

抗生素组合,可实现染色体基因的一步式靶向诱变
细菌染色体的靶向修饰对于了解新药物靶标,研究毒力因子,阐明细胞生理学以及验证基于-组学方法的结果是必要的。对于某些细菌,逆向遗传学仍然是研究进展的主要瓶颈。在这里,我们描述了一种以化合物为中心的策略,该策略将新的阴性选择标记与已知的阳性选择标记结合在一起,以实现简单,高效的细菌染色体一步基因组工程。该方法的灵感来自观察到某些非必需的代谢途径包含必需的晚期步骤,这表明靶向晚期步骤的抗生素可用于选择不存在控制通向该途径的通量的基因。在这个假设的指导下,金黄色葡萄球菌和鲍曼不动杆菌。对于金黄色葡萄球菌,我们使用壁壁磷酸生物合成抑制剂选择不存在tarO,对于鲍曼不动杆菌,我们使用大粘菌素选择不存在lpxC。我们已经在一个单一的平板步骤中以理想的效率获得了所需的基因缺失,基因融合和启动子交换。我们的方法也可以适用于使用FLP重组酶产生基因的无标记缺失。此处描述的工具将加速对两种重要病原体的研究,并且我们概述的概念可以很容易地应用于可以确定合适靶标途径的任何生物。
更新日期:2018-03-13
中文翻译:

抗生素组合,可实现染色体基因的一步式靶向诱变
细菌染色体的靶向修饰对于了解新药物靶标,研究毒力因子,阐明细胞生理学以及验证基于-组学方法的结果是必要的。对于某些细菌,逆向遗传学仍然是研究进展的主要瓶颈。在这里,我们描述了一种以化合物为中心的策略,该策略将新的阴性选择标记与已知的阳性选择标记结合在一起,以实现简单,高效的细菌染色体一步基因组工程。该方法的灵感来自观察到某些非必需的代谢途径包含必需的晚期步骤,这表明靶向晚期步骤的抗生素可用于选择不存在控制通向该途径的通量的基因。在这个假设的指导下,金黄色葡萄球菌和鲍曼不动杆菌。对于金黄色葡萄球菌,我们使用壁壁磷酸生物合成抑制剂选择不存在tarO,对于鲍曼不动杆菌,我们使用大粘菌素选择不存在lpxC。我们已经在一个单一的平板步骤中以理想的效率获得了所需的基因缺失,基因融合和启动子交换。我们的方法也可以适用于使用FLP重组酶产生基因的无标记缺失。此处描述的工具将加速对两种重要病原体的研究,并且我们概述的概念可以很容易地应用于可以确定合适靶标途径的任何生物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号