当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Single-Molecule Titration in a Protein Nanoreactor Reveals the Protonation/Deprotonation Mechanism of a C:C Mismatch in DNA
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2018-03-21 , DOI: 10.1021/jacs.8b00593 Hang Ren 1 , Cameron G. Cheyne 1 , Aaron M. Fleming 1 , Cynthia J. Burrows 1 , Henry S. White 1
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2018-03-21 , DOI: 10.1021/jacs.8b00593 Hang Ren 1 , Cameron G. Cheyne 1 , Aaron M. Fleming 1 , Cynthia J. Burrows 1 , Henry S. White 1
Affiliation
|
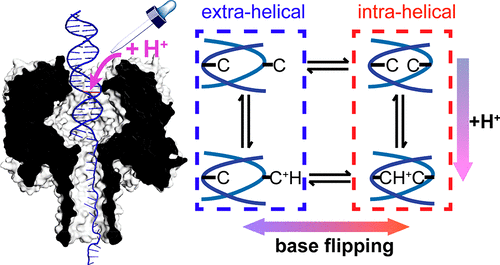
Measurement of single-molecule reactions can elucidate microscopic mechanisms that are often hidden from ensemble analysis. Herein, we report the acid-base titration of a single DNA duplex confined within the wild-type α-hemolysin (α-HL) nanopore for up to 3 h, while monitoring the ionic current through the nanopore. Modulation between two states in the current-time trace for duplexes containing the C:C mismatch in proximity to the latch constriction of α-HL is attributed to the base flipping of the C:C mismatch. As the pH is lowered, the rate for the C:C mismatch to flip from the intra-helical state to the extra-helical state ( kintra-extra) decreases, while the rate for base flipping from the extra-helical state to the intra-helical state ( kextra-intra) remains unchanged. Both kintra-extra and kextra-intra are on the order of 1 × 10-2 s-1 to 1 × 10-1 s-1 and remain stable over the time scale of the measurement (several hours). Analysis of the pH-dependent kinetics of base flipping using a hidden Markov kinetic model demonstrates that protonation/deprotonation occurs while the base pair is in the intra-helical state. We also demonstrate that the rate of protonation is limited by transport of H+ into the α-HL nanopore. Single-molecule kinetic isotope experiments exhibit a large kinetic isotope effect (KIE) for kintra-extra ( kH/ kD ≈ 5) but a limited KIE for kextra-intra ( kH/ kD ≈ 1.3), supporting our model. Our experiments correspond to the longest single-molecule measurements performed using a nanopore, and demonstrate its application in interrogating mechanisms of single-molecule reactions in confined geometries.
中文翻译:

蛋白质纳米反应器中的单分子滴定揭示 DNA 中 C:C 错配的质子化/去质子化机制
单分子反应的测量可以阐明通常隐藏在集合分析中的微观机制。在此,我们报告了限制在野生型 α-溶血素 (α-HL) 纳米孔内的单个 DNA 双链体的酸碱滴定长达 3 小时,同时监测通过纳米孔的离子电流。对于包含 C:C 失配接近 α-HL 锁存收缩的双工,当前时间轨迹中两个状态之间的调制归因于 C:C 失配的基极翻转。随着 pH 值降低,C:C 错配从内螺旋状态翻转到外螺旋状态 (kintra-extra) 的速率降低,而碱基从外螺旋状态翻转到内螺旋状态的速率-螺旋状态(kextra-intra)保持不变。kintra-extra 和 kextra-intra 都在 1 × 10-2 s-1 到 1 × 10-1 s-1 的数量级上,并且在测量的时间尺度(几个小时)内保持稳定。使用隐马尔可夫动力学模型分析碱基翻转的 pH 依赖性动力学表明,当碱基对处于螺旋内状态时,会发生质子化/去质子化。我们还证明质子化率受到 H+ 运输到 α-HL 纳米孔的限制。单分子动力学同位素实验对 kintra-extra ( kH/ kD ≈ 5) 表现出较大的动力学同位素效应 (KIE),但对 kextra-intra ( kH/ kD ≈ 1.3) 表现出有限的 KIE,支持我们的模型。我们的实验对应于使用纳米孔进行的最长单分子测量,并证明了其在研究受限几何结构中单分子反应机制中的应用。
更新日期:2018-03-21
中文翻译:

蛋白质纳米反应器中的单分子滴定揭示 DNA 中 C:C 错配的质子化/去质子化机制
单分子反应的测量可以阐明通常隐藏在集合分析中的微观机制。在此,我们报告了限制在野生型 α-溶血素 (α-HL) 纳米孔内的单个 DNA 双链体的酸碱滴定长达 3 小时,同时监测通过纳米孔的离子电流。对于包含 C:C 失配接近 α-HL 锁存收缩的双工,当前时间轨迹中两个状态之间的调制归因于 C:C 失配的基极翻转。随着 pH 值降低,C:C 错配从内螺旋状态翻转到外螺旋状态 (kintra-extra) 的速率降低,而碱基从外螺旋状态翻转到内螺旋状态的速率-螺旋状态(kextra-intra)保持不变。kintra-extra 和 kextra-intra 都在 1 × 10-2 s-1 到 1 × 10-1 s-1 的数量级上,并且在测量的时间尺度(几个小时)内保持稳定。使用隐马尔可夫动力学模型分析碱基翻转的 pH 依赖性动力学表明,当碱基对处于螺旋内状态时,会发生质子化/去质子化。我们还证明质子化率受到 H+ 运输到 α-HL 纳米孔的限制。单分子动力学同位素实验对 kintra-extra ( kH/ kD ≈ 5) 表现出较大的动力学同位素效应 (KIE),但对 kextra-intra ( kH/ kD ≈ 1.3) 表现出有限的 KIE,支持我们的模型。我们的实验对应于使用纳米孔进行的最长单分子测量,并证明了其在研究受限几何结构中单分子反应机制中的应用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号