当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Binding Specificities of Nanobody•Membrane Protein Complexes Obtained from Chemical Cross-Linking and High-Mass MALDI Mass Spectrometry
Analytical Chemistry ( IF 6.7 ) Pub Date : 2018-03-21 00:00:00 , DOI: 10.1021/acs.analchem.8b00236 Martin Köhler 1 , Christoph Neff 1 , Camilo Perez 2 , Cyrill Brunner 1 , Els Pardon , Jan Steyaert , Gisbert Schneider 1 , Kaspar P. Locher 2 , Renato Zenobi 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2018-03-21 00:00:00 , DOI: 10.1021/acs.analchem.8b00236 Martin Köhler 1 , Christoph Neff 1 , Camilo Perez 2 , Cyrill Brunner 1 , Els Pardon , Jan Steyaert , Gisbert Schneider 1 , Kaspar P. Locher 2 , Renato Zenobi 1
Affiliation
|
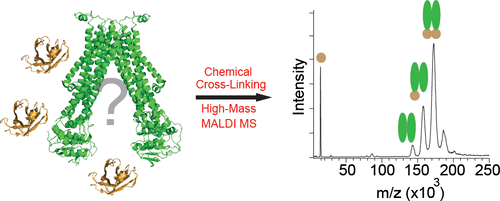
The application of nanobodies as binding partners for structure stabilization in protein X-ray crystallography is taking an increasingly important role in structural biology. However, the addition of nanobodies to the crystallization matrices might complicate the optimization of the crystallization process, which is why analytical techniques to screen and characterize suitable nanobodies are useful. Here, we show how chemical cross-linking combined with high-mass matrix-assisted laser/desorption ionization mass spectrometry can be employed as a fast screening technique to determine binding specificities of intact nanobody•membrane protein complexes. Titration series were performed to rank the binding affinity of the interacting nanobodies. To validate the mass spectrometry data, microscale thermophoresis was used, which showed binding affinities of the stronger binding nanobodies, in the low μM range. In addition, mass spectrometry provides access to the stoichiometry of the complexes formed, which enables the definition of conditions under which homogeneous complex states are present in solution. Conformational changes of the membrane protein were investigated and competitive binding experiments were used to delimit the interaction sites of the nanobodies, which is in agreement with crystal structures obtained. The results show the diversity of specifically binding nanobodies in terms of binding affinity, stoichiometry, and binding site, which illustrates the need for an analytical screening approach.
中文翻译:

化学交联和高质量MALDI质谱法获得的纳米抗体•膜蛋白复合物的结合特异性
纳米抗体作为蛋白质X射线晶体学中结构稳定化的结合伴侣的应用在结构生物学中正发挥着越来越重要的作用。但是,将纳米体添加到结晶基质中可能会使结晶过程的优化复杂化,这就是为什么筛选和表征合适纳米体的分析技术很有用的原因。在这里,我们展示了如何将化学交联与高质量基质辅助激光/解吸电离质谱结合使用,作为确定完整纳米抗体•膜蛋白复合物结合特异性的快速筛选技术。进行滴定系列以对相互作用的纳米抗体的结合亲和力进行排名。为了验证质谱数据,使用了微尺度热泳,在较低的μM范围内,显示出较强结合纳米抗体的结合亲和力。另外,质谱法提供了对所形成的络合物的化学计量的途径,这使得能够定义在溶液中存在均质络合物状态的条件。研究了膜蛋白的构象变化,并通过竞争结合实验确定了纳米抗体的相互作用位点,这与所获得的晶体结构一致。结果显示出在结合亲和力,化学计量和结合位点方面特异性结合的纳米抗体的多样性,这说明了对分析筛选方法的需求。这使得能够定义在溶液中存在均质复杂状态的条件。研究了膜蛋白的构象变化,并通过竞争结合实验确定了纳米抗体的相互作用位点,这与所获得的晶体结构一致。结果显示出在结合亲和力,化学计量和结合位点方面特异性结合的纳米抗体的多样性,这说明了对分析筛选方法的需求。这使得能够定义在溶液中存在均质复杂状态的条件。研究了膜蛋白的构象变化,并通过竞争结合实验确定了纳米抗体的相互作用位点,这与所获得的晶体结构一致。结果显示了在结合亲和力,化学计量和结合位点方面特异性结合的纳米抗体的多样性,这说明了对分析筛选方法的需求。
更新日期:2018-03-21
中文翻译:

化学交联和高质量MALDI质谱法获得的纳米抗体•膜蛋白复合物的结合特异性
纳米抗体作为蛋白质X射线晶体学中结构稳定化的结合伴侣的应用在结构生物学中正发挥着越来越重要的作用。但是,将纳米体添加到结晶基质中可能会使结晶过程的优化复杂化,这就是为什么筛选和表征合适纳米体的分析技术很有用的原因。在这里,我们展示了如何将化学交联与高质量基质辅助激光/解吸电离质谱结合使用,作为确定完整纳米抗体•膜蛋白复合物结合特异性的快速筛选技术。进行滴定系列以对相互作用的纳米抗体的结合亲和力进行排名。为了验证质谱数据,使用了微尺度热泳,在较低的μM范围内,显示出较强结合纳米抗体的结合亲和力。另外,质谱法提供了对所形成的络合物的化学计量的途径,这使得能够定义在溶液中存在均质络合物状态的条件。研究了膜蛋白的构象变化,并通过竞争结合实验确定了纳米抗体的相互作用位点,这与所获得的晶体结构一致。结果显示出在结合亲和力,化学计量和结合位点方面特异性结合的纳米抗体的多样性,这说明了对分析筛选方法的需求。这使得能够定义在溶液中存在均质复杂状态的条件。研究了膜蛋白的构象变化,并通过竞争结合实验确定了纳米抗体的相互作用位点,这与所获得的晶体结构一致。结果显示出在结合亲和力,化学计量和结合位点方面特异性结合的纳米抗体的多样性,这说明了对分析筛选方法的需求。这使得能够定义在溶液中存在均质复杂状态的条件。研究了膜蛋白的构象变化,并通过竞争结合实验确定了纳米抗体的相互作用位点,这与所获得的晶体结构一致。结果显示了在结合亲和力,化学计量和结合位点方面特异性结合的纳米抗体的多样性,这说明了对分析筛选方法的需求。










































 京公网安备 11010802027423号
京公网安备 11010802027423号