Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Kitcre knock-in mice fail to fate-map cardiac stem cells
Nature ( IF 50.5 ) Pub Date : 2018-03-01 , DOI: 10.1038/nature25771 Carla Vicinanza , Iolanda Aquila , Eleonora Cianflone , Mariangela Scalise , Fabiola Marino , Teresa Mancuso , Francesca Fumagalli , Emilia Dora Giovannone , Francesca Cristiano , Enrico Iaccino , Pina Marotta , Annalaura Torella , Roberto Latini , Valter Agosti , Pierangelo Veltri , Konrad Urbanek , Andrea M. Isidori , Dieter Saur , Ciro Indolfi , Bernardo Nadal-Ginard , Daniele Torella
Nature ( IF 50.5 ) Pub Date : 2018-03-01 , DOI: 10.1038/nature25771 Carla Vicinanza , Iolanda Aquila , Eleonora Cianflone , Mariangela Scalise , Fabiola Marino , Teresa Mancuso , Francesca Fumagalli , Emilia Dora Giovannone , Francesca Cristiano , Enrico Iaccino , Pina Marotta , Annalaura Torella , Roberto Latini , Valter Agosti , Pierangelo Veltri , Konrad Urbanek , Andrea M. Isidori , Dieter Saur , Ciro Indolfi , Bernardo Nadal-Ginard , Daniele Torella
|
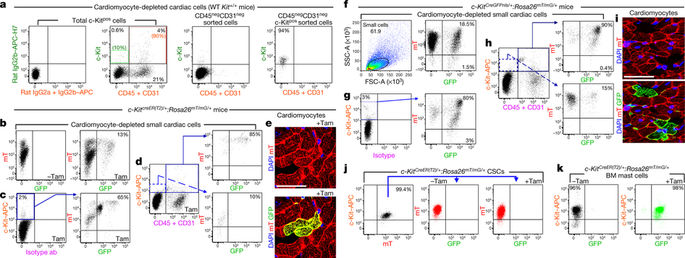
In a cell fate-mapping study1 using a cre-knock-in (KI) into the Kit locus, and in two other studies that used a similar genetic approach to track the fate of cardiac stem/progenitor cells (CSCs)2,3, the authors concluded that c-Kitpos (also known as Kitpos) cells only negligibly contributed to the generation of cardiomyocytes. These studies questioned our findings4 that tissue-specific c-Kitpos CSCs are endogenous regenerative agents that are necessary and sufficient for cardiomyocyte regeneration/replenishment after injury. There is a Reply to this Comment by van Berlo, J. H. et al. Nature 555, http://doi.org/10.1038/ nature25772 (2018). For these differences to be resolved, it is necessary to confirm that the Kitcre-KI approach1 correctly identifies and fate-maps c-Kitpos CSCs and/or investigate whether the insertion of Kitcre-KI affects CSC biology and cardiomyogenic potential. We used tamoxifen-inducible KitCreER(T2)/+(5–7), KitMerCreMer/+ (hereafter KitMCM/+)1,2 and constitutive KitCreGFPnls/+ (ref. 1) mouse lines, which are phenotypically similar to KitW/+ mice5. These mice have white spotting in their fur coat, a 50% decrease in c-Kit expression (Extended Data Fig. 1a) and a testis growth deficit (with low fertility) when heterozygous, this genotype shows fetal/postnatal lethality in homozygotes1–3,6–8. The efficiency of recombination by the Cre–loxP system is proportional to the level of Cre expression and the duration of Cre expression in each cell9–11, which in these Kitcre-KI lines depends on the endogenous Kit promoter. In mice, different c-Kitpos cell types express different levels of c-Kit, with mast cells being the highest c-Kit-expressing cells5–7. In the adult mouse heart, the majority of c-Kitpos cells (≥90%) are committed to the blood cell lineage, expressing markers such as CD45 and CD31 (and are lineage positive (Linpos)) (Fig. 1a). Only a minority (<10%) of c-Kitpos cardiac cells are CD45negCD31neg (Linneg) (Fig. 1a). These Linnegc-Kitpos cardiac cells are enriched for and include all of the adult CSCs, which comprise only approximately 10% of these cells12,13. This subset expresses low but clearly detectable levels of Kit mRNA and c-Kit protein (Extended Data Fig. 1b), which are significantly lower than the levels found in Linposc-Kitpos cardiac cells (Extended Data Fig. 1b). Single wild-type Linnegc-Kitpos clonogenic and multipotent CSCs show significantly lower Kit expression, at the mRNA and protein levels, than embryonic stem cells, haematopoietic stem cells (HSCs) and bone marrow mast cells (Extended Data Fig. 1c, d). Accordingly, CreER(T2) protein and mRNA expression in bone marrow mast cells is robust, whereas it is only faintly detectable in freshly isolated Linneg CSCs (Extended Data Fig. 1e, f). This heterogeneity in cell types that have different levels of c-Kit expression, the low abundance of CSCs among the c-Kitpos cardiac cells and the very low levels of c-Kit expression in CSCs highlight the difficulties of using Kitcre-KI to track the fate of the CSCs. KitCreER(T2)/+ mice6–8 were crossed with homozygous global doublefluorescent Cre-reporter Rosa26mT/mG mice14 (Extended Data Fig. 2a), expressing a membrane-targeted tandem Tomato dimer (mT) that switches to a membrane-targeted GFP (mG) after Cre-dependent recombination14. When double-mutant KitCreER(T2)/+:Rosa26mT/mG/+ mice were given a standard tamoxifen diet for 14 days, 80 ± 8% of the c-Kitpos bone marrow mast cells showed Cre-dependent recombination and expressed GFP, whereas less than 5% of HSCs showed recombination and GFP expression (Extended Data Fig. 1g–j). At the same time, ≤20% of all c-Kitpos cardiac cells were recombined (Extended Data Fig. 1k), but all of these were CD45pos, CD31pos or both, representing cardiac mast cells or endothelial progenitor cells (Extended Data Fig. 1l). By contrast, the CSC-enriched Linnegc-Kitlow cells showed mini mal recombination (≤1%) and expression of GFP (Extended Data Fig. 1l). Using the KitMCM/+:Rosa26mT/mG/+ mice from van Berlo et al.1 yielded almost identical results (Extended Data Fig. 3a–c). To increase the probability of recombination by extending the duration of Cre expression, 8-week-old double-mutant KitCreER(T2)/+: Rosa26mT/mG/+ mice received a tamoxifen diet for four months1 (Fig. 1b–e). This regime showed efficient recombination in several tissue-specific c-Kit-expressing cells (Extended Data Fig. 2b). In the bone marrow, approximately 80% of the total number of cells and up to 60% of c-Kitpos cells became GFPpos (Extended Data Fig. 2c). Analysis of bone marrow cell sub-populations showed that approximately 100% of c-Kitpos mast cells were GFPpos (Extended Data Fig. 2d), whereas only ≤35% of HSCs expressed GFP (Extended Data Fig. 2e). As expected, recombination events were increased further by constitutive Kitcre expression in KitCreGFPnls/+:Rosa26mT/mG/+ double-heterozygous mice (Extended Data Fig. 2g–j). Fluorescence-activated cell sorting (FACS) of dissociated total cardiac cells showed that around 15% of cells showed Cre-mediated recombination and GFP expression (Fig. 1b). Among the total c-Kit antibodylabelled cells, 65 ± 5% were GFPpos (Fig. 1c). Immunohistochemistry of frozen cardiac sections confirmed that approximately 60% of c-Kit antibody-labelled cells had turned GFPpos (Extended Data Fig. 2f). The majority of these GFPpos cells were still mTpos (Fig. 1b, c), highlighting its slow decay in myocardial tissue14. Nevertheless, 80% of the endothelial/mast cell lineage-committed Linposc-Kitpos cells were GFPpos, whereas only ≤10% of the CSC-enriched Linnegc-Kitlow cardiac cells showed recombination (Fig. 1d). Experiments using KitMCM/+ mice1 yielded similar findings (data not shown). After mice were fed the tamoxifen diet for four months, only 0.04 ± 0.01% cardiomyocytes were GFPpos, and most of those cells were also mTpos (Fig. 1e). van Berlo et al.1 interpreted the presence of double-labelled mTpos and GFPpos cardiomyocytes as the evidence of fusion between a pre-existing Rosa26mT/mG/+ cardiomyocyte and a Kitcre/+cell. However, because myocardial decay of mT is very slow14, mT/mG double-positive cardiomyocytes are most likely the consequence of the very slow decay of mT in randomly and progressively recombined progenitors that differentiate into cardiomyocytes. Similar results were obtained when using 8–12-week-old KitCreGFPnls/+:Rosa26mT/mG/+ double-heterozygous mice (Fig. 1f–i) and KitCreGFPnls/+ mice crossed with Rosa26floxed-stop-tdTomato mice (Extended Data Fig. 3d–f). We sorted and cultured Linnegc-Kitpos CSCs from KitCreER(T2)/+: Rosa26mT/mG/+ double-heterozygous mice in the presence or absence of tamoxifen in vitro. Tamoxifen induced recombination in ≤1% of the Linnegc-Kitpos cells and expression of GFP (Fig. 1j), whereas >95% of bone-marrow-derived mast cells showed recombination within seven days (Fig. 1k). None of the single-cell-derived CSC clones that were generated in medium with tamoxifen at passage (P)4
中文翻译:

Kitcre 敲入小鼠未能绘制心脏干细胞的命运图
在使用 cre-knock-in (KI) 进入 Kit 基因座的细胞命运映射研究 1 中,以及在其他两项使用类似遗传方法追踪心脏干/祖细胞 (CSC) 命运的研究中,作者得出结论,c-Kitpos(也称为 Kitpos)细胞对心肌细胞的生成贡献微乎其微。这些研究质疑我们的研究结果 4,即组织特异性 c-Kitpos CSC 是内源性再生剂,是损伤后心肌细胞再生/补充所必需和充分的。van Berlo、JH 等人对此评论进行了回复。Nature 555,http://doi.org/10.1038/nature25772 (2018)。为了解决这些分歧,有必要确认 Kitcre-KI 方法 1 正确识别和命运映射 c-Kitpos CSC 和/或调查 Kitcre-KI 的插入是否影响 CSC 生物学和心肌生成潜力。我们使用他莫昔芬诱导的 KitCreER(T2)/+(5–7)、KitMerCreMer/+(以下称为 KitMCM/+)1,2 和组成型 KitCreGFPnls/+(参考文献 1)小鼠系,它们在表型上与 KitW/+ 相似老鼠 5. 这些小鼠的皮毛上有白点,c-Kit 表达降低 50%(扩展数据图 1a),杂合子时睾丸生长缺陷(生育力低),这种基因型显示纯合子的胎儿/产后致死率 1-3 ,6–8。Cre-loxP 系统的重组效率与每个细胞中 Cre 表达的水平和 Cre 表达的持续时间成正比9-11,在这些 Kitcre-KI 品系中,这取决于内源性 Kit 启动子。在小鼠中,不同的 c-Kitpos 细胞类型表达不同水平的 c-Kit,肥大细胞是最高的 c-Kit 表达细胞 5-7。在成年小鼠心脏中,大多数 c-Kitpos 细胞(≥90%)都属于血细胞谱系,表达 CD45 和 CD31 等标记物(并且谱系阳性(Linpos))(图 1a)。只有少数 (<10%) c-Kitpos 心脏细胞是 CD45negCD31neg (Linneg)(图 1a)。这些 Linnegc-Kitpos 心脏细胞富含并包括所有成体 CSC,这些细胞仅占这些细胞的 12,13 的大约 10%。该子集表达低但清晰可检测的 Kit mRNA 和 c-Kit 蛋白水平(扩展数据图 1b),其显着低于在 Linposc-Kitpos 心脏细胞中发现的水平(扩展数据图 1b)。与胚胎干细胞、造血干细胞 (HSC) 和骨髓肥大细胞相比,单个野生型 Linnegc-Kitpos 克隆形成和多能 CSC 在 mRNA 和蛋白质水平上显示出显着较低的 Kit 表达(扩展数据图 1c、d)。因此,骨髓肥大细胞中的 CreER(T2) 蛋白和 mRNA 表达很强,而在新鲜分离的 Linneg CSC 中只能微弱地检测到(扩展数据图 1e,f)。具有不同 c-Kit 表达水平的细胞类型的这种异质性、c-Kitpos 心脏细胞中 CSC 的丰度低以及 CSC 中 c-Kit 的表达水平非常低,这凸显了使用 Kitcre-KI 来追踪CSC 的命运。KitCreER(T2)/+ 小鼠 6-8 与纯合的全局双荧光 Cre 报告基因 Rosa26mT/mG 小鼠杂交 14(扩展数据图 2a),表达膜靶向串联番茄二聚体 (mT),在 Cre 依赖性重组后转换为膜靶向 GFP (mG)。当给双突变 KitCreER(T2)/+:Rosa26mT/mG/+ 小鼠标准他莫昔芬饮食 14 天时,80 ± 8% 的 c-Kitpos 骨髓肥大细胞表现出依赖 Cre 的重组并表达 GFP,而不到 5% 的 HSC 显示重组和 GFP 表达(扩展数据图 1g-j)。同时,≤20%的所有c-Kitpos心肌细胞被重组(扩展数据图1k),但所有这些都是CD45pos、CD31pos或两者,代表心脏肥大细胞或内皮祖细胞(扩展数据图1l) )。相比之下,富含 CSC 的 Linnegc-Kitlow 细胞显示出最小的重组(≤1%)和 GFP 的表达(扩展数据图 1l)。使用 KitMCM/+:来自 van Berlo 等人 1 的 Rosa26mT/mG/+ 小鼠产生了几乎相同的结果(扩展数据图 3a-c)。为了通过延长 Cre 表达的持续时间来增加重组的可能性,8 周大的双突变 KitCreER(T2)/+: Rosa26mT/mG/+ 小鼠接受了他莫昔芬饮食四个月1(图 1b-e)。该方案在几种组织特异性 c-Kit 表达细胞中显示出有效的重组(扩展数据图 2b)。在骨髓中,大约 80% 的细胞总数和高达 60% 的 c-Kitpos 细胞变成了 GFPpos(扩展数据图 2c)。骨髓细胞亚群的分析表明,大约 100% 的 c-Kitpos 肥大细胞是 GFPpos(扩展数据图 2d),而只有≤35% 的 HSC 表达 GFP(扩展数据图 2e)。正如预期的那样,KitCreGFPnls/+:Rosa26mT/mG/+ 双杂合小鼠中的组成型 Kitcre 表达进一步增加了重组事件(扩展数据图 2g-j)。分离的总心脏细胞的荧光激活细胞分选(FACS)显示大约 15% 的细胞显示 Cre 介导的重组和 GFP 表达(图 1b)。在总 c-Kit 抗体标记的细胞中,65±5% 是 GFPpos(图 1c)。冷冻心脏切片的免疫组织化学证实,大约 60% 的 c-Kit 抗体标记细胞已转变为 GFPpos(扩展数据图 2f)。大多数这些 GFPpos 细胞仍然是 mTpos(图 1b、c),突出了其在心肌组织中的缓慢衰减 14。尽管如此,80% 的内皮/肥大细胞谱系定向的 Linposc-Kitpos 细胞是 GFPpos,而只有≤10% 的富含 CSC 的 Linnegc-Kitlow 心脏细胞显示重组(图 1d)。使用 KitMCM/+ 小鼠 1 的实验产生了类似的结果(数据未显示)。在小鼠喂食他莫昔芬饮食四个月后,只有 0.04 ± 0.01% 的心肌细胞是 GFPpos,而这些细胞中的大部分也是 mTpos(图 1e)。van Berlo 等人 1 将双标记的 mTpos 和 GFPpos 心肌细胞的存在解释为预先存在的 Rosa26mT/mG/+ 心肌细胞和 Kitcre/+ 细胞之间融合的证据。然而,由于 mT 的心肌衰减非常缓慢 14,mT/mG 双阳性心肌细胞很可能是随机和渐进重组的祖细胞中 mT 非常缓慢衰减的结果,这些祖细胞分化为心肌细胞。使用 8-12 周龄 KitCreGFPnls/+ 时获得了类似的结果:Rosa26mT/mG/+ 双杂合小鼠(图 1f-i)和 KitCreGFPnls/+ 小鼠与 Rosa26floxed-stop-tdTomato 小鼠杂交(扩展数据图 3d-f)。我们在体外在存在或不存在他莫昔芬的情况下,从 KitCreER(T2)/+: Rosa26mT/mG/+ 双杂合小鼠中分选并培养了 Linnegc-Kitpos CSC。他莫昔芬在≤1%的Linnegc-Kitpos细胞中诱导重组和GFP表达(图1j),而>95%的骨髓来源肥大细胞在7天内显示重组(图1k)。在传代 (P)4 时在含有他莫昔芬的培养基中产生的单细胞衍生的 CSC 克隆中没有一个 Rosa26mT/mG/+ 双杂合小鼠在体外存在或不存在他莫昔芬的情况。他莫昔芬在≤1%的Linnegc-Kitpos细胞中诱导重组和GFP表达(图1j),而>95%的骨髓来源肥大细胞在7天内显示重组(图1k)。在传代 (P)4 时,在含有他莫昔芬的培养基中产生的单细胞衍生 CSC 克隆中没有一个 Rosa26mT/mG/+ 双杂合小鼠在体外存在或不存在他莫昔芬的情况。他莫昔芬在≤1%的Linnegc-Kitpos细胞中诱导重组和GFP表达(图1j),而>95%的骨髓来源肥大细胞在7天内显示重组(图1k)。在传代 (P)4 时,在含有他莫昔芬的培养基中产生的单细胞衍生 CSC 克隆中没有一个
更新日期:2018-03-01
中文翻译:

Kitcre 敲入小鼠未能绘制心脏干细胞的命运图
在使用 cre-knock-in (KI) 进入 Kit 基因座的细胞命运映射研究 1 中,以及在其他两项使用类似遗传方法追踪心脏干/祖细胞 (CSC) 命运的研究中,作者得出结论,c-Kitpos(也称为 Kitpos)细胞对心肌细胞的生成贡献微乎其微。这些研究质疑我们的研究结果 4,即组织特异性 c-Kitpos CSC 是内源性再生剂,是损伤后心肌细胞再生/补充所必需和充分的。van Berlo、JH 等人对此评论进行了回复。Nature 555,http://doi.org/10.1038/nature25772 (2018)。为了解决这些分歧,有必要确认 Kitcre-KI 方法 1 正确识别和命运映射 c-Kitpos CSC 和/或调查 Kitcre-KI 的插入是否影响 CSC 生物学和心肌生成潜力。我们使用他莫昔芬诱导的 KitCreER(T2)/+(5–7)、KitMerCreMer/+(以下称为 KitMCM/+)1,2 和组成型 KitCreGFPnls/+(参考文献 1)小鼠系,它们在表型上与 KitW/+ 相似老鼠 5. 这些小鼠的皮毛上有白点,c-Kit 表达降低 50%(扩展数据图 1a),杂合子时睾丸生长缺陷(生育力低),这种基因型显示纯合子的胎儿/产后致死率 1-3 ,6–8。Cre-loxP 系统的重组效率与每个细胞中 Cre 表达的水平和 Cre 表达的持续时间成正比9-11,在这些 Kitcre-KI 品系中,这取决于内源性 Kit 启动子。在小鼠中,不同的 c-Kitpos 细胞类型表达不同水平的 c-Kit,肥大细胞是最高的 c-Kit 表达细胞 5-7。在成年小鼠心脏中,大多数 c-Kitpos 细胞(≥90%)都属于血细胞谱系,表达 CD45 和 CD31 等标记物(并且谱系阳性(Linpos))(图 1a)。只有少数 (<10%) c-Kitpos 心脏细胞是 CD45negCD31neg (Linneg)(图 1a)。这些 Linnegc-Kitpos 心脏细胞富含并包括所有成体 CSC,这些细胞仅占这些细胞的 12,13 的大约 10%。该子集表达低但清晰可检测的 Kit mRNA 和 c-Kit 蛋白水平(扩展数据图 1b),其显着低于在 Linposc-Kitpos 心脏细胞中发现的水平(扩展数据图 1b)。与胚胎干细胞、造血干细胞 (HSC) 和骨髓肥大细胞相比,单个野生型 Linnegc-Kitpos 克隆形成和多能 CSC 在 mRNA 和蛋白质水平上显示出显着较低的 Kit 表达(扩展数据图 1c、d)。因此,骨髓肥大细胞中的 CreER(T2) 蛋白和 mRNA 表达很强,而在新鲜分离的 Linneg CSC 中只能微弱地检测到(扩展数据图 1e,f)。具有不同 c-Kit 表达水平的细胞类型的这种异质性、c-Kitpos 心脏细胞中 CSC 的丰度低以及 CSC 中 c-Kit 的表达水平非常低,这凸显了使用 Kitcre-KI 来追踪CSC 的命运。KitCreER(T2)/+ 小鼠 6-8 与纯合的全局双荧光 Cre 报告基因 Rosa26mT/mG 小鼠杂交 14(扩展数据图 2a),表达膜靶向串联番茄二聚体 (mT),在 Cre 依赖性重组后转换为膜靶向 GFP (mG)。当给双突变 KitCreER(T2)/+:Rosa26mT/mG/+ 小鼠标准他莫昔芬饮食 14 天时,80 ± 8% 的 c-Kitpos 骨髓肥大细胞表现出依赖 Cre 的重组并表达 GFP,而不到 5% 的 HSC 显示重组和 GFP 表达(扩展数据图 1g-j)。同时,≤20%的所有c-Kitpos心肌细胞被重组(扩展数据图1k),但所有这些都是CD45pos、CD31pos或两者,代表心脏肥大细胞或内皮祖细胞(扩展数据图1l) )。相比之下,富含 CSC 的 Linnegc-Kitlow 细胞显示出最小的重组(≤1%)和 GFP 的表达(扩展数据图 1l)。使用 KitMCM/+:来自 van Berlo 等人 1 的 Rosa26mT/mG/+ 小鼠产生了几乎相同的结果(扩展数据图 3a-c)。为了通过延长 Cre 表达的持续时间来增加重组的可能性,8 周大的双突变 KitCreER(T2)/+: Rosa26mT/mG/+ 小鼠接受了他莫昔芬饮食四个月1(图 1b-e)。该方案在几种组织特异性 c-Kit 表达细胞中显示出有效的重组(扩展数据图 2b)。在骨髓中,大约 80% 的细胞总数和高达 60% 的 c-Kitpos 细胞变成了 GFPpos(扩展数据图 2c)。骨髓细胞亚群的分析表明,大约 100% 的 c-Kitpos 肥大细胞是 GFPpos(扩展数据图 2d),而只有≤35% 的 HSC 表达 GFP(扩展数据图 2e)。正如预期的那样,KitCreGFPnls/+:Rosa26mT/mG/+ 双杂合小鼠中的组成型 Kitcre 表达进一步增加了重组事件(扩展数据图 2g-j)。分离的总心脏细胞的荧光激活细胞分选(FACS)显示大约 15% 的细胞显示 Cre 介导的重组和 GFP 表达(图 1b)。在总 c-Kit 抗体标记的细胞中,65±5% 是 GFPpos(图 1c)。冷冻心脏切片的免疫组织化学证实,大约 60% 的 c-Kit 抗体标记细胞已转变为 GFPpos(扩展数据图 2f)。大多数这些 GFPpos 细胞仍然是 mTpos(图 1b、c),突出了其在心肌组织中的缓慢衰减 14。尽管如此,80% 的内皮/肥大细胞谱系定向的 Linposc-Kitpos 细胞是 GFPpos,而只有≤10% 的富含 CSC 的 Linnegc-Kitlow 心脏细胞显示重组(图 1d)。使用 KitMCM/+ 小鼠 1 的实验产生了类似的结果(数据未显示)。在小鼠喂食他莫昔芬饮食四个月后,只有 0.04 ± 0.01% 的心肌细胞是 GFPpos,而这些细胞中的大部分也是 mTpos(图 1e)。van Berlo 等人 1 将双标记的 mTpos 和 GFPpos 心肌细胞的存在解释为预先存在的 Rosa26mT/mG/+ 心肌细胞和 Kitcre/+ 细胞之间融合的证据。然而,由于 mT 的心肌衰减非常缓慢 14,mT/mG 双阳性心肌细胞很可能是随机和渐进重组的祖细胞中 mT 非常缓慢衰减的结果,这些祖细胞分化为心肌细胞。使用 8-12 周龄 KitCreGFPnls/+ 时获得了类似的结果:Rosa26mT/mG/+ 双杂合小鼠(图 1f-i)和 KitCreGFPnls/+ 小鼠与 Rosa26floxed-stop-tdTomato 小鼠杂交(扩展数据图 3d-f)。我们在体外在存在或不存在他莫昔芬的情况下,从 KitCreER(T2)/+: Rosa26mT/mG/+ 双杂合小鼠中分选并培养了 Linnegc-Kitpos CSC。他莫昔芬在≤1%的Linnegc-Kitpos细胞中诱导重组和GFP表达(图1j),而>95%的骨髓来源肥大细胞在7天内显示重组(图1k)。在传代 (P)4 时在含有他莫昔芬的培养基中产生的单细胞衍生的 CSC 克隆中没有一个 Rosa26mT/mG/+ 双杂合小鼠在体外存在或不存在他莫昔芬的情况。他莫昔芬在≤1%的Linnegc-Kitpos细胞中诱导重组和GFP表达(图1j),而>95%的骨髓来源肥大细胞在7天内显示重组(图1k)。在传代 (P)4 时,在含有他莫昔芬的培养基中产生的单细胞衍生 CSC 克隆中没有一个 Rosa26mT/mG/+ 双杂合小鼠在体外存在或不存在他莫昔芬的情况。他莫昔芬在≤1%的Linnegc-Kitpos细胞中诱导重组和GFP表达(图1j),而>95%的骨髓来源肥大细胞在7天内显示重组(图1k)。在传代 (P)4 时,在含有他莫昔芬的培养基中产生的单细胞衍生 CSC 克隆中没有一个











































 京公网安备 11010802027423号
京公网安备 11010802027423号