当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Small Molecule Interactome Mapping by Photoaffinity Labeling Reveals Binding Site Hotspots for the NSAIDs
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2018-03-15 , DOI: 10.1021/jacs.7b11639 Jinxu Gao 1 , Adelphe Mfuh 1 , Yuka Amako 1 , Christina M. Woo 1
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2018-03-15 , DOI: 10.1021/jacs.7b11639 Jinxu Gao 1 , Adelphe Mfuh 1 , Yuka Amako 1 , Christina M. Woo 1
Affiliation
|
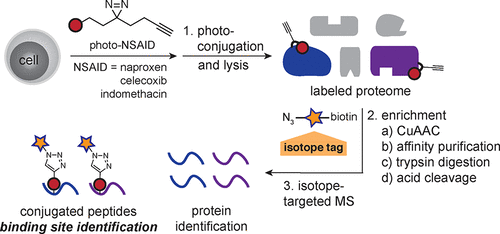
Many therapeutics elicit cell-type specific polypharmacology that is executed by a network of molecular recognition events between a small molecule and the whole proteome. However, measurement of the structures that underpin the molecular associations between the proteome and even common therapeutics, such as the nonsteroidal anti-inflammatory drugs (NSAIDs), is limited by the inability to map the small molecule interactome. To address this gap, we developed a platform termed small molecule interactome mapping by photoaffinity labeling (SIM-PAL) and applied it to the in cellulo direct characterization of specific NSAID binding sites. SIM-PAL uses (1) photochemical conjugation of NSAID derivatives in the whole proteome and (2) enrichment and isotope-recoding of the conjugated peptides for (3) targeted mass spectrometry-based assignment. Using SIM-PAL, we identified the NSAID interactome consisting of over 1000 significantly enriched proteins and directly characterized nearly 200 conjugated peptides representing direct binding sites of the photo-NSAIDs with proteins from Jurkat and K562 cells. The enriched proteins were often identified as parts of complexes, including known targets of NSAID activity (e.g., NF-κB) and novel interactions (e.g., AP-2, proteasome). The conjugated peptides revealed direct NSAID binding sites from the cell surface to the nucleus and a specific binding site hotspot for the three photo-NSAIDs on histones H2A and H2B. NSAID binding stabilized COX-2 and histone H2A by cellular thermal shift assay. Since small molecule stabilization of protein complexes is a gain of function regulatory mechanism, it is conceivable that NSAIDs affect biological processes through these broader proteomic interactions. SIM-PAL enabled characterization of NSAID binding site hotspots and is amenable to map global binding sites for virtually any molecule of interest.
中文翻译:

光亲和标记的小分子相互作用组图谱揭示了 NSAID 的结合位点热点
许多疗法引发细胞类型特异性多药理学,由小分子和整个蛋白质组之间的分子识别事件网络执行。然而,由于无法绘制小分子相互作用组,因此无法测量蛋白质组和甚至常见疗法(例如非甾体抗炎药 (NSAID))之间的分子关联的结构。为了解决这一差距,我们开发了一个称为通过光亲和标记 (SIM-PAL) 进行小分子相互作用组映射的平台,并将其应用于特定 NSAID 结合位点的纤维素内直接表征。SIM-PAL 使用 (1) NSAID 衍生物在整个蛋白质组中的光化学偶联和 (2) 偶联肽的富集和同位素记录,用于 (3) 基于质谱的靶向分配。使用 SIM-PAL,我们鉴定了由 1000 多种显着富集的蛋白质组成的 NSAID 相互作用组,并直接表征了近 200 种偶联肽,这些偶联肽代表了来自 Jurkat 和 K562 细胞的蛋白质的 photo-NSAID 的直接结合位点。富集的蛋白质通常被鉴定为复合物的一部分,包括 NSAID 活性的已知靶标(例如,NF-κB)和新的相互作用(例如,AP-2、蛋白酶体)。缀合的肽揭示了从细胞表面到细胞核的直接 NSAID 结合位点,以及组蛋白 H2A 和 H2B 上三种光-NSAID 的特异性结合位点热点。通过细胞热位移测定,NSAID 结合稳定了 COX-2 和组蛋白 H2A。由于蛋白质复合物的小分子稳定是功能调节机制的获得,可以想象,NSAIDs 通过这些更广泛的蛋白质组学相互作用影响生物过程。SIM-PAL 能够表征 NSAID 结合位点热点,并且能够绘制几乎任何感兴趣分子的全局结合位点。
更新日期:2018-03-15
中文翻译:

光亲和标记的小分子相互作用组图谱揭示了 NSAID 的结合位点热点
许多疗法引发细胞类型特异性多药理学,由小分子和整个蛋白质组之间的分子识别事件网络执行。然而,由于无法绘制小分子相互作用组,因此无法测量蛋白质组和甚至常见疗法(例如非甾体抗炎药 (NSAID))之间的分子关联的结构。为了解决这一差距,我们开发了一个称为通过光亲和标记 (SIM-PAL) 进行小分子相互作用组映射的平台,并将其应用于特定 NSAID 结合位点的纤维素内直接表征。SIM-PAL 使用 (1) NSAID 衍生物在整个蛋白质组中的光化学偶联和 (2) 偶联肽的富集和同位素记录,用于 (3) 基于质谱的靶向分配。使用 SIM-PAL,我们鉴定了由 1000 多种显着富集的蛋白质组成的 NSAID 相互作用组,并直接表征了近 200 种偶联肽,这些偶联肽代表了来自 Jurkat 和 K562 细胞的蛋白质的 photo-NSAID 的直接结合位点。富集的蛋白质通常被鉴定为复合物的一部分,包括 NSAID 活性的已知靶标(例如,NF-κB)和新的相互作用(例如,AP-2、蛋白酶体)。缀合的肽揭示了从细胞表面到细胞核的直接 NSAID 结合位点,以及组蛋白 H2A 和 H2B 上三种光-NSAID 的特异性结合位点热点。通过细胞热位移测定,NSAID 结合稳定了 COX-2 和组蛋白 H2A。由于蛋白质复合物的小分子稳定是功能调节机制的获得,可以想象,NSAIDs 通过这些更广泛的蛋白质组学相互作用影响生物过程。SIM-PAL 能够表征 NSAID 结合位点热点,并且能够绘制几乎任何感兴趣分子的全局结合位点。



























 京公网安备 11010802027423号
京公网安备 11010802027423号