Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The cis-regulatory dynamics of embryonic development at single-cell resolution
Nature ( IF 50.5 ) Pub Date : 2018-03-01 , DOI: 10.1038/nature25981 Darren A Cusanovich 1 , James P Reddington 2 , David A Garfield 2 , Riza M Daza 1 , Delasa Aghamirzaie 1 , Raquel Marco-Ferreres 2 , Hannah A Pliner 1 , Lena Christiansen 3 , Xiaojie Qiu 1 , Frank J Steemers 3 , Cole Trapnell 1 , Jay Shendure 1, 4 , Eileen E M Furlong 2
Nature ( IF 50.5 ) Pub Date : 2018-03-01 , DOI: 10.1038/nature25981 Darren A Cusanovich 1 , James P Reddington 2 , David A Garfield 2 , Riza M Daza 1 , Delasa Aghamirzaie 1 , Raquel Marco-Ferreres 2 , Hannah A Pliner 1 , Lena Christiansen 3 , Xiaojie Qiu 1 , Frank J Steemers 3 , Cole Trapnell 1 , Jay Shendure 1, 4 , Eileen E M Furlong 2
Affiliation
|
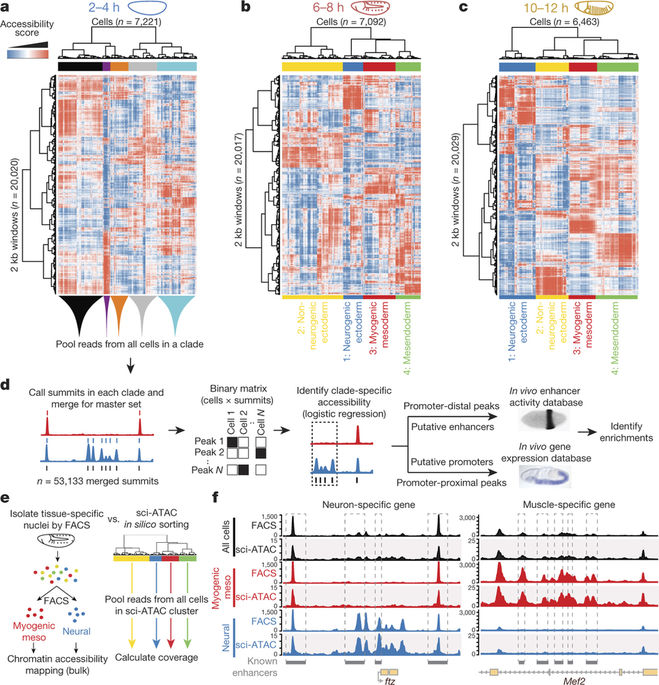
Understanding how gene regulatory networks control the progressive restriction of cell fates is a long-standing challenge. Recent advances in measuring gene expression in single cells are providing new insights into lineage commitment. However, the regulatory events underlying these changes remain unclear. Here we investigate the dynamics of chromatin regulatory landscapes during embryogenesis at single-cell resolution. Using single-cell combinatorial indexing assay for transposase accessible chromatin with sequencing (sci-ATAC-seq), we profiled chromatin accessibility in over 20,000 single nuclei from fixed Drosophila melanogaster embryos spanning three landmark embryonic stages: 2–4 h after egg laying (predominantly stage 5 blastoderm nuclei), when each embryo comprises around 6,000 multipotent cells; 6–8 h after egg laying (predominantly stage 10–11), to capture a midpoint in embryonic development when major lineages in the mesoderm and ectoderm are specified; and 10–12 h after egg laying (predominantly stage 13), when each of the embryo’s more than 20,000 cells are undergoing terminal differentiation. Our results show that there is spatial heterogeneity in the accessibility of the regulatory genome before gastrulation, a feature that aligns with future cell fate, and that nuclei can be temporally ordered along developmental trajectories. During mid-embryogenesis, tissue granularity emerges such that individual cell types can be inferred by their chromatin accessibility while maintaining a signature of their germ layer of origin. Analysis of the data reveals overlapping usage of regulatory elements between cells of the endoderm and non-myogenic mesoderm, suggesting a common developmental program that is reminiscent of the mesendoderm lineage in other species. We identify 30,075 distal regulatory elements that exhibit tissue-specific accessibility. We validated the germ-layer specificity of a subset of these predicted enhancers in transgenic embryos, achieving an accuracy of 90%. Overall, our results demonstrate the power of shotgun single-cell profiling of embryos to resolve dynamic changes in the chromatin landscape during development, and to uncover the cis-regulatory programs of metazoan germ layers and cell types.
中文翻译:

单细胞分辨率下胚胎发育的顺式调控动力学
了解基因调控网络如何控制细胞命运的渐进限制是一个长期存在的挑战。测量单细胞基因表达的最新进展为谱系定型提供了新的见解。然而,这些变化背后的监管事件仍不清楚。在这里,我们以单细胞分辨率研究胚胎发生过程中染色质调控景观的动态。使用转座酶可及染色质的单细胞组合索引分析和测序 (sci-ATAC-seq),我们分析了来自固定果蝇胚胎的 20,000 多个单核的染色质可及性,跨越三个标志性胚胎阶段:产卵后 2-4 小时(主要是产卵后 2-4 小时)阶段 5 胚盘核),此时每个胚胎包含约 6,000 个专能细胞;产蛋后 6-8 小时(主要是第 10-11 阶段),以捕获胚胎发育的中点,此时中胚层和外胚层的主要谱系已明确;产卵后 10-12 小时(主要是第 13 阶段),此时每个胚胎的 20,000 多个细胞正在进行终末分化。我们的结果表明,原肠胚形成前调控基因组的可及性存在空间异质性,这一特征与未来的细胞命运相一致,并且细胞核可以沿着发育轨迹在时间上排序。在胚胎发生中期,组织粒度出现,使得单个细胞类型可以通过其染色质可及性来推断,同时保持其胚层起源的特征。对数据的分析揭示了内胚层和非肌源性中胚层细胞之间调节元件的重叠使用,这表明了一个共同的发育程序,让人想起其他物种的中内胚层谱系。 我们确定了 30,075 个具有组织特异性可及性的远端调控元件。我们验证了转基因胚胎中这些预测增强子的子集的胚层特异性,准确率达到 90%。总的来说,我们的结果证明了胚胎鸟枪法单细胞分析的能力,可以解决发育过程中染色质景观的动态变化,并揭示后生动物胚层和细胞类型的顺式调控程序。
更新日期:2018-03-01
中文翻译:

单细胞分辨率下胚胎发育的顺式调控动力学
了解基因调控网络如何控制细胞命运的渐进限制是一个长期存在的挑战。测量单细胞基因表达的最新进展为谱系定型提供了新的见解。然而,这些变化背后的监管事件仍不清楚。在这里,我们以单细胞分辨率研究胚胎发生过程中染色质调控景观的动态。使用转座酶可及染色质的单细胞组合索引分析和测序 (sci-ATAC-seq),我们分析了来自固定果蝇胚胎的 20,000 多个单核的染色质可及性,跨越三个标志性胚胎阶段:产卵后 2-4 小时(主要是产卵后 2-4 小时)阶段 5 胚盘核),此时每个胚胎包含约 6,000 个专能细胞;产蛋后 6-8 小时(主要是第 10-11 阶段),以捕获胚胎发育的中点,此时中胚层和外胚层的主要谱系已明确;产卵后 10-12 小时(主要是第 13 阶段),此时每个胚胎的 20,000 多个细胞正在进行终末分化。我们的结果表明,原肠胚形成前调控基因组的可及性存在空间异质性,这一特征与未来的细胞命运相一致,并且细胞核可以沿着发育轨迹在时间上排序。在胚胎发生中期,组织粒度出现,使得单个细胞类型可以通过其染色质可及性来推断,同时保持其胚层起源的特征。对数据的分析揭示了内胚层和非肌源性中胚层细胞之间调节元件的重叠使用,这表明了一个共同的发育程序,让人想起其他物种的中内胚层谱系。 我们确定了 30,075 个具有组织特异性可及性的远端调控元件。我们验证了转基因胚胎中这些预测增强子的子集的胚层特异性,准确率达到 90%。总的来说,我们的结果证明了胚胎鸟枪法单细胞分析的能力,可以解决发育过程中染色质景观的动态变化,并揭示后生动物胚层和细胞类型的顺式调控程序。









































 京公网安备 11010802027423号
京公网安备 11010802027423号