Talanta ( IF 5.6 ) Pub Date : 2018-03-08 , DOI: 10.1016/j.talanta.2018.02.109 Laetitia Minh Maï Le , Balázs Kégl , Alexandre Gramfort , Camille Marini , David Nguyen , Mehdi Cherti , Sana Tfaili , Ali Tfayli , Arlette Baillet-Guffroy , Patrice Prognon , Pierre Chaminade , Eric Caudron
|
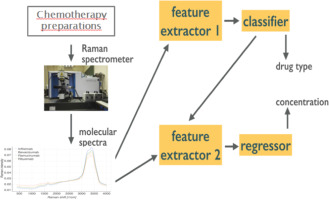
The use of monoclonal antibodies (mAbs) constitutes one of the most important strategies to treat patients suffering from cancers such as hematological malignancies and solid tumors. These antibodies are prescribed by the physician and prepared by hospital pharmacists. An analytical control enables the quality of the preparations to be ensured. The aim of this study was to explore the development of a rapid analytical method for quality control. The method used four mAbs (Infliximab, Bevacizumab, Rituximab and Ramucirumab) at various concentrations and was based on recording Raman data and coupling them to a traditional chemometric and machine learning approach for data analysis. Compared to conventional linear approach, prediction errors are reduced with a data-driven approach using statistical machine learning methods. In the latter, preprocessing and predictive models are jointly optimized. An additional original aspect of the work involved on submitting the problem to a collaborative data challenge platform called Rapid Analytics and Model Prototyping (RAMP). This allowed using solutions from about 300 data scientists in collaborative work. Using machine learning, the prediction of the four mAbs samples was considerably improved. The best predictive model showed a combined error of 2.4% versus 14.6% using linear approach. The concentration and classification errors were 5.8% and 0.7%, only three spectra were misclassified over the 429 spectra of the test set. This large improvement obtained with machine learning techniques was uniform for all molecules but maximal for Bevacizumab with an 88.3% reduction on combined errors (2.1% versus 17.9%).
中文翻译:

使用协作机器学习方法优化拉曼光谱中四种单克隆抗体的分类和回归分析
单克隆抗体(mAb)的使用构成了治疗患有癌症(如血液系统恶性肿瘤和实体瘤)的患者的最重要策略之一。这些抗体由医师开具处方,并由医院药剂师制备。分析控制可确保制剂的质量。这项研究的目的是探索质量控制快速分析方法的发展。该方法使用了各种浓度的四种mAb(英夫利昔单抗,贝伐单抗,利妥昔单抗和雷米库单抗),其基础是记录拉曼数据,并将其与传统的化学计量学和机器学习方法耦合以进行数据分析。与传统的线性方法相比,使用统计机器学习方法的数据驱动方法可减少预测误差。在后者中,联合优化了预处理和预测模型。该工作的另一个原始方面涉及将问题提交给称为Rapid Analytics and Model Prototyping(RAMP)的协作数据挑战平台。这允许在协作工作中使用大约300名数据科学家的解决方案。使用机器学习,对四个mAb样品的预测有了显着改善。最佳预测模型的综合误差为2.4%对比使用线性方法的14.6%浓度和分类误差分别为5.8%和0.7%,在测试集的429个光谱中只有三个光谱被错误分类。通过机器学习技术获得的这一大进步对于所有分子都是统一的,但对于贝伐单抗而言却是最大的,合并错误减少了88.3%(2.1%对17.9%)。











































 京公网安备 11010802027423号
京公网安备 11010802027423号