当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Rapid and low-cost strategy for detecting genome-editing induced deletion: A single-copy case
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2018-08-01 , DOI: 10.1016/j.aca.2018.02.060 Nan Cheng , Qin Wang , Ying Shang , Yuancong Xu , Kunlun Huang , Zhansen Yang , Dengke Pan , Wentao Xu , Yunbo Luo
Analytica Chimica Acta ( IF 6.2 ) Pub Date : 2018-08-01 , DOI: 10.1016/j.aca.2018.02.060 Nan Cheng , Qin Wang , Ying Shang , Yuancong Xu , Kunlun Huang , Zhansen Yang , Dengke Pan , Wentao Xu , Yunbo Luo
|
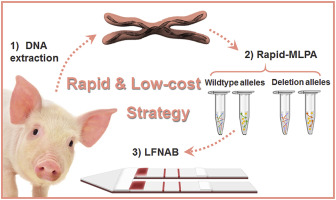
Genome editing techniques have been implemented in human daily lives, which has created a high demand for the development of new gene-edited product analysis methods. Conventional assays are time-consuming, labor-intensive, and costly. This paper proposes a rapid and low-cost strategy for detecting genome-editing induced deletion which works by integrating rapid-multiplex ligation-dependent probe amplification (MLPA) with a dual-lateral flow nucleic acid biosensor (LFNAB) cascade in a single-copy case. A rapid-MLPA was first introduced to the LFNAB system as a replacement for the conventional PCR for enhanced specificity and accuracy. A dual-LFNAB was applied for the detection of genome-editing induced deletion without any additional instrumentation or complex operation. After optimization, we achieved the specific detection of wildtype alleles and deletion alleles in spiked samples with a detection limit of 0.4 fM, which is comparable to that of electrophoresis-based detection assays and fluorescent biosensors. To confirm the validity and feasibility of our strategy, we assayed two pork samples from two WUZHISHAN pigs successfully. By comparing the detection results from next-generation sequencing analysis, we found that the proposed cascade demonstrates at least 20-fold shorter assay time and at least 100-fold less assay cost. To this effect, the proposed method is a rapid and low-cost solution to sample-to-answer detection of genome-editing induced deletion and shows remarkable potential in regards to international trade, transparency, and freedom of choice.
中文翻译:

检测基因组编辑诱导缺失的快速低成本策略:单拷贝案例
基因组编辑技术已经在人类日常生活中得到应用,这对开发新的基因编辑产品分析方法产生了很高的需求。传统检测费时、费力且成本高昂。本文提出了一种快速、低成本的检测基因组编辑诱导缺失的策略,该策略通过将快速多重连接依赖性探针扩增 (MLPA) 与双侧流核酸生物传感器 (LFNAB) 级联整合到单拷贝中来发挥作用。案件。LFNAB 系统首先引入了快速 MLPA,作为传统 PCR 的替代品,以提高特异性和准确性。双 LFNAB 用于检测基因组编辑诱导的缺失,无需任何额外的仪器或复杂的操作。优化后,我们实现了对加标样品中野生型等位基因和缺失等位基因的特异性检测,检测限为 0.4 fM,与基于电泳的检测分析和荧光生物传感器的检测限相当。为了证实我们的策略的有效性和可行性,我们成功地检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。这与基于电泳的检测分析和荧光生物传感器相当。为了证实我们的策略的有效性和可行性,我们成功地检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。这与基于电泳的检测分析和荧光生物传感器相当。为了证实我们的策略的有效性和可行性,我们成功地检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。我们成功检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。我们成功检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。
更新日期:2018-08-01
中文翻译:

检测基因组编辑诱导缺失的快速低成本策略:单拷贝案例
基因组编辑技术已经在人类日常生活中得到应用,这对开发新的基因编辑产品分析方法产生了很高的需求。传统检测费时、费力且成本高昂。本文提出了一种快速、低成本的检测基因组编辑诱导缺失的策略,该策略通过将快速多重连接依赖性探针扩增 (MLPA) 与双侧流核酸生物传感器 (LFNAB) 级联整合到单拷贝中来发挥作用。案件。LFNAB 系统首先引入了快速 MLPA,作为传统 PCR 的替代品,以提高特异性和准确性。双 LFNAB 用于检测基因组编辑诱导的缺失,无需任何额外的仪器或复杂的操作。优化后,我们实现了对加标样品中野生型等位基因和缺失等位基因的特异性检测,检测限为 0.4 fM,与基于电泳的检测分析和荧光生物传感器的检测限相当。为了证实我们的策略的有效性和可行性,我们成功地检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。这与基于电泳的检测分析和荧光生物传感器相当。为了证实我们的策略的有效性和可行性,我们成功地检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。这与基于电泳的检测分析和荧光生物传感器相当。为了证实我们的策略的有效性和可行性,我们成功地检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。我们成功检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。我们成功检测了两只五指山猪的两个猪肉样品。通过比较下一代测序分析的检测结果,我们发现所提出的级联证明至少缩短了 20 倍的检测时间,并且至少降低了 100 倍的检测成本。为此,所提出的方法是一种快速、低成本的解决方案,用于基因组编辑诱导缺失的样本到答案检测,并在国际贸易、透明度和选择自由方面显示出显着的潜力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号