Journal of Allergy and Clinical Immunology ( IF 14.2 ) Pub Date : 2018-03-05 , DOI: 10.1016/j.jaci.2018.02.020 Mina Fazlollahi , Tricia D. Lee , Jade Andrade , Kasopefoluwa Oguntuyo , Yoojin Chun , Galina Grishina , Alexander Grishin , Supinda Bunyavanich
|
Background
Nasal microbiota may influence asthma pathobiology.
Objective
We sought to characterize the nasal microbiome of subjects with exacerbated asthma, nonexacerbated asthma, and healthy controls to identify nasal microbiota associated with asthma activity.
Methods
We performed 16S ribosomal RNA sequencing on nasal swabs obtained from 72 primarily adult subjects with exacerbated asthma (n = 20), nonexacerbated asthma (n = 31), and healthy controls (n = 21). Analyses were performed using Quantitative Insights into Microbial (QIIME); linear discriminant analysis effect size (LEfSe); Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; and Statistical Analysis of Metagenomic Profiles (PICRUSt); and Statistical Analysis of Metagenomic Profiles (STAMP). Species found to be associated with asthma activity were validated using quantitative PCR. Metabolic pathways associated with differentially abundant nasal taxa were inferred through metagenomic functional prediction.
Results
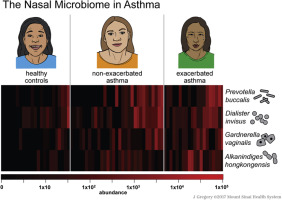
Nasal bacterial composition significantly differed among subjects with exacerbated asthma, nonexacerbated asthma, and healthy controls (permutational multivariate ANOVA, P = 2.2 × 10−2). Relative to controls, the nasal microbiota of subjects with asthma were enriched with taxa from Bacteroidetes (Wilcoxon-Mann-Whitney, r = 0.33, P = 5.1 × 10−3) and Proteobacteria (r = 0.29, P = 1.4 × 10−2). Four species were differentially abundant based on asthma status after correction for multiple comparisons: Prevotella buccalis, Padj = 1.0 × 10−2; Dialister invisus, Padj = 9.1 × 10−3; Gardnerella vaginalis, Padj = 2.8 × 10−3; Alkanindiges hongkongensis, Padj = 2.6 × 10−3. These phyla and species were also differentially abundant based on asthma activity (exacerbated asthma vs nonexacerbated asthma vs controls). Quantitative PCR confirmed species overrepresentation in asthma relative to controls for Prevotella buccalis (fold change = 130, P = 2.1 × 10−4) and Gardnerella vaginalis (fold change = 160, P = 6.8 × 10−4). Metagenomic inference revealed differential glycerolipid metabolism (Kruskal-Wallis, P = 1.9 × 10−4) based on asthma activity.
Conclusions
Nasal microbiome composition differs in subjects with exacerbated asthma, nonexacerbated asthma, and healthy controls. The identified nasal taxa could be further investigated for potential mechanistic roles in asthma and as possible biomarkers of asthma activity.



























 京公网安备 11010802027423号
京公网安备 11010802027423号