当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Cover Image, Volume 39, Issue 11
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2018-03-01 , DOI: 10.1002/jcc.25203
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2018-03-01 , DOI: 10.1002/jcc.25203

|
In the cover article on page 621, Trung Hai Nguyen and co‐authors calculated protein‐ligand interaction energies at points along a 3D grid using fast Fourier transform. These interaction energies are employed in binding free energy calculations. The cover image is based on an ‐11 kcal/mol isosurface and evenly‐spaced volume slices of the interaction energy grid. The secondary structure of the protein, T4 lysozyme, is shown as a mauve ribbon. The ligand, benzene, is placed in the binding pocket and shown in a licorice representation. (DOI: 10.1002/jcc.25139)
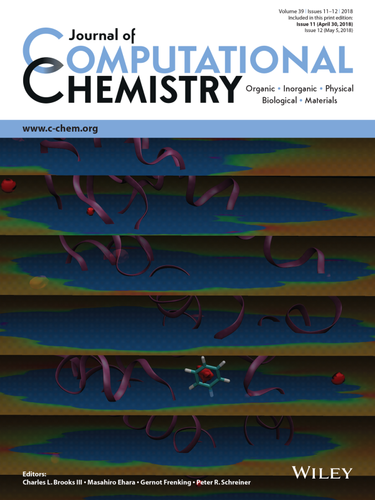
中文翻译:

封面图片,第39卷,第11期
在第621页的封面文章中,Trung Hai Nguyen及其合作者使用快速傅立叶变换计算了沿3D网格的点处的蛋白质-配体相互作用能。这些相互作用能被用于结合自由能的计算。封面图像基于-11 kcal / mol的等值面和相互作用能网格的均匀间隔的体积切片。蛋白质的二级结构T4溶菌酶显示为淡紫色带。配体苯置于结合袋中,并以甘草表示。(DOI:10.1002 / jcc.25139)

更新日期:2018-03-01

中文翻译:

封面图片,第39卷,第11期
在第621页的封面文章中,Trung Hai Nguyen及其合作者使用快速傅立叶变换计算了沿3D网格的点处的蛋白质-配体相互作用能。这些相互作用能被用于结合自由能的计算。封面图像基于-11 kcal / mol的等值面和相互作用能网格的均匀间隔的体积切片。蛋白质的二级结构T4溶菌酶显示为淡紫色带。配体苯置于结合袋中,并以甘草表示。(DOI:10.1002 / jcc.25139)




























 京公网安备 11010802027423号
京公网安备 11010802027423号