当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Comprehensive Analysis of Anion–Quadrupole Interactions in Protein Structures
Biochemistry ( IF 2.9 ) Pub Date : 2018-02-26 00:00:00 , DOI: 10.1021/acs.biochem.7b01006 Suvobrata Chakravarty 1, 2 , Adron R Ung 1 , Brian Moore 3 , Jay Shore 1 , Mona Alshamrani 1
Biochemistry ( IF 2.9 ) Pub Date : 2018-02-26 00:00:00 , DOI: 10.1021/acs.biochem.7b01006 Suvobrata Chakravarty 1, 2 , Adron R Ung 1 , Brian Moore 3 , Jay Shore 1 , Mona Alshamrani 1
Affiliation
|
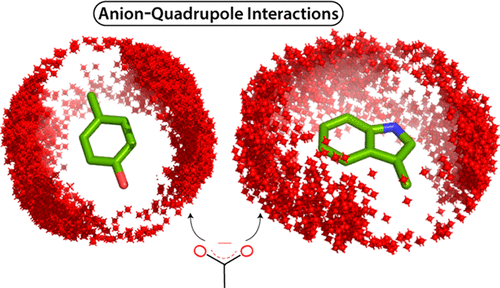
The edgewise interactions of anions with phenylalanine (Phe) aromatic rings in proteins, known as anion–quadrupole interactions, have been well studied. However, the anion–quadrupole interactions of the tyrosine (Tyr) and tryptophan (Trp) rings have been less well studied, probably because these have been considered weaker than interactions of anions hydrogen bonded to Trp/Tyr side chains. Distinguishing such hydrogen bonding interactions, we comprehensively surveyed the edgewise interactions of certain anions (aspartate, glutamate, and phosphate) with Trp, Tyr, and Phe rings in high-resolution, nonredundant protein single chains and interfaces (protein–protein, DNA/RNA–protein, and membrane–protein). Trp/Tyr anion–quadrupole interactions are common, with Trp showing the highest propensity and average interaction energy for this type of interaction. The energy of an anion–quadrupole interaction (−15.0 to 0.0 kcal/mol, based on quantum mechanical calculations) depends not only on the interaction geometry but also on the ring atom. The phosphate anions at DNA/RNA–protein interfaces interact with aromatic residues with energies comparable to that of aspartate/glutamate anion–quadrupole interactions. At DNA–protein interfaces, the frequency of aromatic ring participation in anion–quadrupole interactions is comparable to that of positive charge participation in salt bridges, suggesting an underappreciated role for anion–quadrupole interactions at DNA–protein (or membrane–protein) interfaces. Although less frequent than salt bridges in single-chain proteins, we observed highly conserved anion–quadrupole interactions in the structures of remote homologues, and evolutionary covariance-based residue contact score predictions suggest that conserved anion–quadrupole interacting pairs, like salt bridges, contribute to polypeptide folding, stability, and recognition.
中文翻译:

蛋白质结构中阴离子-四极相互作用的综合分析
阴离子与蛋白质中苯丙氨酸 (Phe) 芳环的边缘相互作用(称为阴离子四极相互作用)已得到充分研究。然而,酪氨酸 (Tyr) 和色氨酸 (Trp) 环的阴离子-四极相互作用研究较少,可能是因为这些相互作用被认为弱于与 Trp/Tyr 侧链氢键结合的阴离子相互作用。为了区分这种氢键相互作用,我们全面研究了高分辨率、非冗余蛋白质单链和界面(蛋白质-蛋白质、DNA/RNA)中某些阴离子(天冬氨酸盐、谷氨酸盐和磷酸盐)与 Trp、Tyr 和 Phe 环的边缘相互作用。 –蛋白质和膜蛋白质)。 Trp/Tyr 阴离子-四极相互作用很常见,其中 Trp 显示出此类相互作用的最高倾向和平均相互作用能。阴离子-四极相互作用的能量(-15.0 至 0.0 kcal/mol,基于量子力学计算)不仅取决于相互作用的几何形状,还取决于环原子。 DNA/RNA-蛋白质界面处的磷酸根阴离子与芳香族残基相互作用,其能量与天冬氨酸/谷氨酸阴离子-四极相互作用的能量相当。在DNA-蛋白质界面,芳香环参与阴离子-四极相互作用的频率与正电荷参与盐桥的频率相当,这表明阴离子-四极相互作用在DNA-蛋白质(或膜-蛋白质)界面中的作用未被充分认识。 尽管在单链蛋白中频率低于盐桥,但我们在远程同源物的结构中观察到高度保守的阴离子-四极相互作用,并且基于进化协方差的残基接触分数预测表明,保守的阴离子-四极相互作用对,如盐桥,有助于多肽折叠、稳定性和识别。
更新日期:2018-02-26
中文翻译:

蛋白质结构中阴离子-四极相互作用的综合分析
阴离子与蛋白质中苯丙氨酸 (Phe) 芳环的边缘相互作用(称为阴离子四极相互作用)已得到充分研究。然而,酪氨酸 (Tyr) 和色氨酸 (Trp) 环的阴离子-四极相互作用研究较少,可能是因为这些相互作用被认为弱于与 Trp/Tyr 侧链氢键结合的阴离子相互作用。为了区分这种氢键相互作用,我们全面研究了高分辨率、非冗余蛋白质单链和界面(蛋白质-蛋白质、DNA/RNA)中某些阴离子(天冬氨酸盐、谷氨酸盐和磷酸盐)与 Trp、Tyr 和 Phe 环的边缘相互作用。 –蛋白质和膜蛋白质)。 Trp/Tyr 阴离子-四极相互作用很常见,其中 Trp 显示出此类相互作用的最高倾向和平均相互作用能。阴离子-四极相互作用的能量(-15.0 至 0.0 kcal/mol,基于量子力学计算)不仅取决于相互作用的几何形状,还取决于环原子。 DNA/RNA-蛋白质界面处的磷酸根阴离子与芳香族残基相互作用,其能量与天冬氨酸/谷氨酸阴离子-四极相互作用的能量相当。在DNA-蛋白质界面,芳香环参与阴离子-四极相互作用的频率与正电荷参与盐桥的频率相当,这表明阴离子-四极相互作用在DNA-蛋白质(或膜-蛋白质)界面中的作用未被充分认识。 尽管在单链蛋白中频率低于盐桥,但我们在远程同源物的结构中观察到高度保守的阴离子-四极相互作用,并且基于进化协方差的残基接触分数预测表明,保守的阴离子-四极相互作用对,如盐桥,有助于多肽折叠、稳定性和识别。










































 京公网安备 11010802027423号
京公网安备 11010802027423号