当前位置:
X-MOL 学术
›
J. Phys. Chem. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Dynamics and Interactions of a 29 kDa Human Enzyme Studied by Solid-State NMR
The Journal of Physical Chemistry Letters ( IF 5.7 ) Pub Date : 2018-02-26 00:00:00 , DOI: 10.1021/acs.jpclett.8b00110 Suresh K. Vasa 1, 2 , Himanshu Singh 1, 2 , Petra Rovó 1, 2 , Rasmus Linser 1, 2
The Journal of Physical Chemistry Letters ( IF 5.7 ) Pub Date : 2018-02-26 00:00:00 , DOI: 10.1021/acs.jpclett.8b00110 Suresh K. Vasa 1, 2 , Himanshu Singh 1, 2 , Petra Rovó 1, 2 , Rasmus Linser 1, 2
Affiliation
|
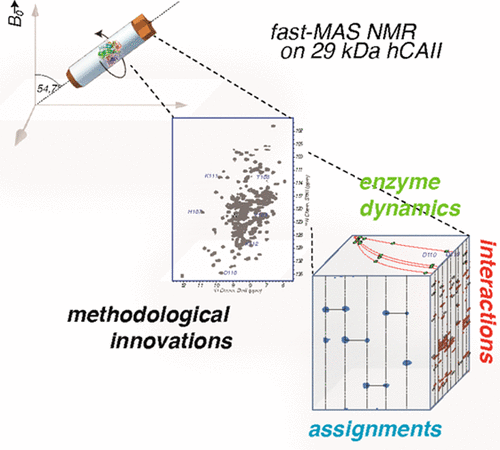
Solid-state NMR has been employed for characterization of a broad range of biomacromolecules and supramolecular assemblies. However, because of limitations in sensitivity and resolution, the size of the individual monomeric units has rarely exceeded 15 kDa. As such, enzymes, which are often more complex and comprise long peptide chains, have not been easily accessible, even though manifold desirable information could potentially be provided by solid-state NMR studies. Here, we demonstrate that more than 1200 backbone and side-chain chemical shifts can be reliably assessed from minimal sample quantities for a 29 kDa human enzyme of the carbonic anhydrase family, giving access to its backbone dynamics and intermolecular interactions with a small-molecule inhibitor. The possibility of comprehensive assessment of enzymes in this molecular-weight regime without molecular-tumbling-derived limitations enables the study of residue-specific properties important for their mode of action as well as for pharmacological interference in this and many other enzymes.
中文翻译:

固态NMR研究29 kDa人类酶的动力学和相互作用
固态NMR已用于表征各种生物大分子和超分子组装体。但是,由于灵敏度和分辨率的限制,单个单体单元的大小很少超过15 kDa。因此,尽管固态NMR研究可能潜在地提供了多种所需信息,但通常更复杂且包含长肽链的酶仍不容易获得。在这里,我们证明了对于碳酸酐酶家族的29 kDa人类酶而言,最少样品量即可可靠地评估1200多个主链和侧链化学位移,从而可利用其与小分子抑制剂的主链动力学和分子间相互作用。
更新日期:2018-02-26
中文翻译:

固态NMR研究29 kDa人类酶的动力学和相互作用
固态NMR已用于表征各种生物大分子和超分子组装体。但是,由于灵敏度和分辨率的限制,单个单体单元的大小很少超过15 kDa。因此,尽管固态NMR研究可能潜在地提供了多种所需信息,但通常更复杂且包含长肽链的酶仍不容易获得。在这里,我们证明了对于碳酸酐酶家族的29 kDa人类酶而言,最少样品量即可可靠地评估1200多个主链和侧链化学位移,从而可利用其与小分子抑制剂的主链动力学和分子间相互作用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号