当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Improved Analysis of RNA Localization by Spatially Restricted Oxidation of RNA–Protein Complexes
Biochemistry ( IF 2.9 ) Pub Date : 2018-02-23 00:00:00 , DOI: 10.1021/acs.biochem.8b00053 Ying Li , Mahima B. Aggarwal , Ke Ke , Kim Nguyen , Robert C. Spitale
Biochemistry ( IF 2.9 ) Pub Date : 2018-02-23 00:00:00 , DOI: 10.1021/acs.biochem.8b00053 Ying Li , Mahima B. Aggarwal , Ke Ke , Kim Nguyen , Robert C. Spitale
|
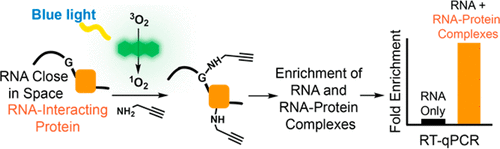
Recent analysis of transcriptomes has revealed that RNAs perform a myriad of functions beyond encoding proteins. Critical to RNA function is its transport to unique subcellular locations. Despite the importance of RNA localization, it is still very challenging to study in an unbiased manner. We recently described the ability to tag RNA molecules within subcellular locations through spatially restricted nucleobase oxidation. Herein, we describe a dramatic improvement of this protocol through the localized oxidation and tagging of proteins. Isolation of RNA–protein complexes enabled the enrichment of challenging RNA targets on chromatin and presented a considerably optimized protocol for the analysis of RNA subcellular localization within living cells.
中文翻译:

通过 RNA-蛋白质复合物的空间限制氧化改进 RNA 定位分析
最近对转录组的分析表明,RNA 除了编码蛋白质之外,还具有多种功能。RNA 功能的关键是将其运输到独特的亚细胞位置。尽管RNA定位很重要,但以公正的方式进行研究仍然非常具有挑战性。我们最近描述了通过空间限制的核碱基氧化来标记亚细胞位置内的 RNA 分子的能力。在这里,我们描述了通过蛋白质的局部氧化和标记对该协议的显着改进。RNA-蛋白质复合物的分离能够富集染色质上具有挑战性的 RNA 靶标,并为活细胞内 RNA 亚细胞定位的分析提供了相当优化的方案。
更新日期:2018-02-23
中文翻译:

通过 RNA-蛋白质复合物的空间限制氧化改进 RNA 定位分析
最近对转录组的分析表明,RNA 除了编码蛋白质之外,还具有多种功能。RNA 功能的关键是将其运输到独特的亚细胞位置。尽管RNA定位很重要,但以公正的方式进行研究仍然非常具有挑战性。我们最近描述了通过空间限制的核碱基氧化来标记亚细胞位置内的 RNA 分子的能力。在这里,我们描述了通过蛋白质的局部氧化和标记对该协议的显着改进。RNA-蛋白质复合物的分离能够富集染色质上具有挑战性的 RNA 靶标,并为活细胞内 RNA 亚细胞定位的分析提供了相当优化的方案。











































 京公网安备 11010802027423号
京公网安备 11010802027423号