当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular Mechanism, Dynamics, and Energetics of Protein-Mediated Dinucleotide Flipping in a Mismatched DNA: A Computational Study of the RAD4-DNA Complex
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-02-23 00:00:00 , DOI: 10.1021/acs.jcim.7b00636 Kartheek Pitta 1 , Marimuthu Krishnan 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2018-02-23 00:00:00 , DOI: 10.1021/acs.jcim.7b00636 Kartheek Pitta 1 , Marimuthu Krishnan 1
Affiliation
|
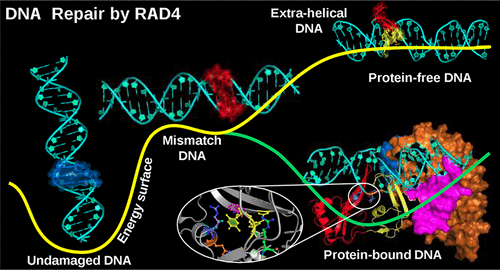
DNA damage alters genetic information and adversely affects gene expression pathways leading to various complex genetic disorders and cancers. DNA repair proteins recognize and rectify DNA damage and mismatches with high fidelity. A critical molecular event that occurs during most protein-mediated DNA repair processes is the extrusion of orphaned bases at the damaged site facilitated by specific repairing enzymes. The molecular-level understanding of the mechanism, dynamics, and energetics of base extrusion is necessary to elucidate the molecular basis of protein-mediated DNA damage repair. The present article investigates the molecular mechanism of dinucleotide extrusion in a mismatched DNA (containing a stretch of three contiguous thymidine–thymidine base pairs) facilitated by Radiation sensitive 4 (RAD4), a key DNA repair protein, on an atom-by-atom basis using molecular dynamics (MD) and umbrella-sampling (US) simulations. Using atomistic models of RAD4-free and RAD4-bound mismatched DNA, the free energy profiles associated with extrusion of mismatched partner bases are determined for both systems. The mismatched bases adopted the most stable intrahelical conformation, and their extrusion was unfavorable in RAD4-free mismatched DNA due to the presence of prohibitively high barriers (>12.0 kcal/mol) along the extrusion pathways. Upon binding of RAD4 to the DNA, the global free energy minimum is shifted to the extrahelical state indicating the key role of RAD4-DNA interactions in catalyzing the dinucleotide base extrusion in the DNA-RAD4 complex. The critical residues of RAD4 contributing to the conformational stability of the mismatched bases are identified, and the energetics of insertion of a β-hairpin of RAD4 into the DNA duplex is examined. The conformational energy landscape-based mechanistic insight into RAD4-mediated base extrusion provided here may serve as a useful baseline to understand the molecular basis of xeroderma pigmentosum C (XPC)-mediated DNA damage repair in humans.
中文翻译:

错配DNA中蛋白质介导的二核苷酸翻转的分子机理,动力学和能量学:RAD4-DNA复合体的计算研究
DNA损伤会改变遗传信息,并对基因表达途径产生不利影响,从而导致各种复杂的遗传性疾病和癌症。DNA修复蛋白可以高保真地识别和纠正DNA损伤和错配。在大多数蛋白质介导的DNA修复过程中发生的关键分子事件是在特定修复酶的帮助下,孤儿碱基在受损部位的挤出。分子水平的基础挤压的机制,动力学和能量学的理解是必要的,以阐明蛋白质介导的DNA损伤修复的分子基础。本文研究了由关键的DNA修复蛋白辐射敏感4(RAD4)促进的错配DNA(包含三个连续的胸腺嘧啶-胸腺嘧啶碱基对的片段)中双核苷酸挤出的分子机制,使用分子动力学(MD)和伞式采样(US)模拟在逐个原子的基础上进行。使用无RAD4和RAD4结合的错配DNA的原子模型,可以确定两个系统与错配伴侣碱基的挤出相关的自由能分布。错配的碱基采用最稳定的螺旋内构象,由于沿挤出途径存在过高的屏障(> 12.0 kcal / mol),因此它们的挤出在不含RAD4的错配DNA中是不利的。在RAD4与DNA结合后,整体的自由能最小值移至螺旋外状态,表明RAD4-DNA相互作用在催化DNA-RAD4复合物中的二核苷酸碱基挤出中起关键作用。鉴定了RAD4的关键残基,这些残基有助于错配碱基的构象稳定性,并且研究了将RAD4的β-发夹插入DNA双链体的能量学。此处提供的有关RAD4介导的基础挤压的基于构象能态景观的机械洞察力,可以作为了解人类干性色素C(XPC)介导的DNA损伤修复的分子基础的有用基线。
更新日期:2018-02-23
中文翻译:

错配DNA中蛋白质介导的二核苷酸翻转的分子机理,动力学和能量学:RAD4-DNA复合体的计算研究
DNA损伤会改变遗传信息,并对基因表达途径产生不利影响,从而导致各种复杂的遗传性疾病和癌症。DNA修复蛋白可以高保真地识别和纠正DNA损伤和错配。在大多数蛋白质介导的DNA修复过程中发生的关键分子事件是在特定修复酶的帮助下,孤儿碱基在受损部位的挤出。分子水平的基础挤压的机制,动力学和能量学的理解是必要的,以阐明蛋白质介导的DNA损伤修复的分子基础。本文研究了由关键的DNA修复蛋白辐射敏感4(RAD4)促进的错配DNA(包含三个连续的胸腺嘧啶-胸腺嘧啶碱基对的片段)中双核苷酸挤出的分子机制,使用分子动力学(MD)和伞式采样(US)模拟在逐个原子的基础上进行。使用无RAD4和RAD4结合的错配DNA的原子模型,可以确定两个系统与错配伴侣碱基的挤出相关的自由能分布。错配的碱基采用最稳定的螺旋内构象,由于沿挤出途径存在过高的屏障(> 12.0 kcal / mol),因此它们的挤出在不含RAD4的错配DNA中是不利的。在RAD4与DNA结合后,整体的自由能最小值移至螺旋外状态,表明RAD4-DNA相互作用在催化DNA-RAD4复合物中的二核苷酸碱基挤出中起关键作用。鉴定了RAD4的关键残基,这些残基有助于错配碱基的构象稳定性,并且研究了将RAD4的β-发夹插入DNA双链体的能量学。此处提供的有关RAD4介导的基础挤压的基于构象能态景观的机械洞察力,可以作为了解人类干性色素C(XPC)介导的DNA损伤修复的分子基础的有用基线。











































 京公网安备 11010802027423号
京公网安备 11010802027423号