当前位置:
X-MOL 学术
›
ACS Infect. Dis.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Using in Vitro Evolution and Whole Genome Analysis To Discover Next Generation Targets for Antimalarial Drug Discovery.
ACS Infectious Diseases ( IF 4.0 ) Pub Date : 2018-02-21 , DOI: 10.1021/acsinfecdis.7b00276 Madeline R Luth 1 , Purva Gupta 1 , Sabine Ottilie 1 , Elizabeth A Winzeler 1, 2
ACS Infectious Diseases ( IF 4.0 ) Pub Date : 2018-02-21 , DOI: 10.1021/acsinfecdis.7b00276 Madeline R Luth 1 , Purva Gupta 1 , Sabine Ottilie 1 , Elizabeth A Winzeler 1, 2
Affiliation
|
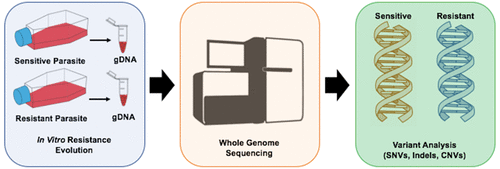
Although many new anti-infectives have been discovered and developed solely using phenotypic cellular screening and assay optimization, most researchers recognize that structure-guided drug design is more practical and less costly. In addition, a greater chemical space can be interrogated with structure-guided drug design. The practicality of structure-guided drug design has launched a search for the targets of compounds discovered in phenotypic screens. One method that has been used extensively in malaria parasites for target discovery and chemical validation is in vitro evolution and whole genome analysis (IVIEWGA). Here, small molecules from phenotypic screens with demonstrated antiparasitic activity are used in genome-based target discovery methods. In this Review, we discuss the newest, most promising druggable targets discovered or further validated by evolution-based methods, as well as some exceptions.
中文翻译:

使用体外进化和全基因组分析发现抗疟疾药物发现的下一代目标。
尽管仅使用表型细胞筛选和分析优化已发现和开发了许多新的抗感染药,但大多数研究人员认识到,结构导向的药物设计更加实用且成本更低。此外,结构导向的药物设计可以询问更大的化学空间。结构指导药物设计的实用性已经开始寻找在表型筛选中发现的化合物的靶标。在疟原虫中广泛用于目标发现和化学验证的一种方法是体外进化和全基因组分析(IVIEWGA)。在这里,来自表型筛选的具有证明的抗寄生虫活性的小分子被用于基于基因组的靶标发现方法中。在这篇评论中,我们讨论了最新的,
更新日期:2018-02-16
中文翻译:

使用体外进化和全基因组分析发现抗疟疾药物发现的下一代目标。
尽管仅使用表型细胞筛选和分析优化已发现和开发了许多新的抗感染药,但大多数研究人员认识到,结构导向的药物设计更加实用且成本更低。此外,结构导向的药物设计可以询问更大的化学空间。结构指导药物设计的实用性已经开始寻找在表型筛选中发现的化合物的靶标。在疟原虫中广泛用于目标发现和化学验证的一种方法是体外进化和全基因组分析(IVIEWGA)。在这里,来自表型筛选的具有证明的抗寄生虫活性的小分子被用于基于基因组的靶标发现方法中。在这篇评论中,我们讨论了最新的,











































 京公网安备 11010802027423号
京公网安备 11010802027423号