Journal of Allergy and Clinical Immunology ( IF 11.4 ) Pub Date : 2018-02-21 , DOI: 10.1016/j.jaci.2017.12.992 Steve Jupe , Keith Ray , Corina Duenas Roca , Thawfeek Varusai , Veronica Shamovsky , Lincoln Stein , Peter D'Eustachio , Henning Hermjakob
|
Background
There is a wealth of biological pathway information available in the scientific literature, but it is spread across many thousands of publications. Alongside publications that contain definitive experimental discoveries are many others that have been dismissed as spurious, found to be irreproducible, or are contradicted by later results and consequently now considered controversial. Many descriptions and images of pathways are incomplete stylized representations that assume the reader is an expert and familiar with the established details of the process, which are consequently not fully explained. Pathway representations in publications frequently do not represent a complete, detailed, and unambiguous description of the molecules involved; their precise posttranslational state; or a full account of the molecular events they undergo while participating in a process. Although this might be sufficient to be interpreted by an expert reader, the lack of detail makes such pathways less useful and difficult to understand for anyone unfamiliar with the area and of limited use as the basis for computational models.
Objective
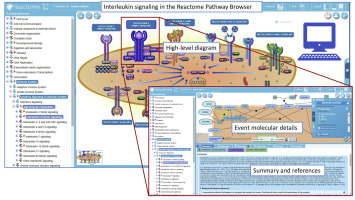
Reactome was established as a freely accessible knowledge base of human biological pathways. It is manually populated with interconnected molecular events that fully detail the molecular participants linked to published experimental data and background material by using a formal and open data structure that facilitates computational reuse. These data are accessible on a Web site in the form of pathway diagrams that have descriptive summaries and annotations and as downloadable data sets in several formats that can be reused with other computational tools. The entire database and all supporting software can be downloaded and reused under a Creative Commons license.
Methods
Pathways are authored by expert biologists who work with Reactome curators and editorial staff to represent the consensus in the field. Pathways are represented as interactive diagrams that include as much molecular detail as possible and are linked to literature citations that contain supporting experimental details. All newly created events undergo a peer-review process before they are added to the database and made available on the associated Web site. New content is added quarterly.
Results
The 63rd release of Reactome in December 2017 contains 10,996 human proteins participating in 11,426 events in 2,179 pathways. In addition, analytic tools allow data set submission for the identification and visualization of pathway enrichment and representation of expression profiles as an overlay on Reactome pathways. Protein-protein and compound-protein interactions from several sources, including custom user data sets, can be added to extend pathways. Pathway diagrams and analytic result displays can be downloaded as editable images, human-readable reports, and files in several standard formats that are suitable for computational reuse. Reactome content is available programmatically through a REpresentational State Transfer (REST)-based content service and as a Neo4J graph database. Signaling pathways for IL-1 to IL-38 are hierarchically classified within the pathway “signaling by interleukins.” The classification used is largely derived from Akdis et al.
Conclusion
The addition to Reactome of a complete set of the known human interleukins, their receptors, and established signaling pathways linked to annotations of relevant aspects of immune function provides a significant computationally accessible resource of information about this important family. This information can be extended easily as new discoveries become accepted as the consensus in the field. A key aim for the future is to increase coverage of gene expression changes induced by interleukin signaling.
中文翻译:

白细胞介素及其信号传导途径在Reactome生物学途径数据库中
背景
科学文献中有大量的生物途径信息,但分布在成千上万的出版物中。除了包含确定性实验发现的出版物外,还有许多其他出版物被认为是虚假的,不可复制的,或者与后来的结果相矛盾,因此现在被认为是有争议的。路径的许多描述和图像都是不完整的程式化表示,它们假定读者是专家并且熟悉该过程的已建立细节,因此未完全说明。出版物中的途径表示常常不能代表所涉及分子的完整,详细和明确的描述;其确切的翻译后状态;或完整说明他们在参与过程中所经历的分子事件。尽管这足以由专业读者解释,但由于缺乏细节,因此对于那些不熟悉该领域并且使用有限的人作为计算模型的基础,此类途径的用处不大,也难以理解。
客观的
Reactome已建立为可免费获取的人类生物学途径的知识库。它使用互连的分子事件进行手动填充,这些事件通过使用正式的开放数据结构(有助于计算重用)来详细描述与已发布的实验数据和背景材料链接的分子参与者。这些数据可以通过路径图的形式在网站上进行访问,这些路径图具有描述性的摘要和注释,并且可以几种格式下载为可下载的数据集,可以与其他计算工具一起使用。整个数据库和所有支持软件都可以在Creative Commons许可下下载并重新使用。
方法
途径是由与Reactome策展人和编辑人员一起工作的专家生物学家撰写的,代表了该领域的共识。通路以交互图表示,其中包括尽可能多的分子细节,并与包含支持性实验细节的文献引文链接。所有新创建的事件在添加到数据库并在关联的网站上可用之前,都经过同行评审过程。新内容每季度添加一次。
结果
2017年12月发布的第63版Reactome包含10,996种人类蛋白质,参与了2,179条途径的11,426个事件。此外,分析工具允许提交数据集,以鉴定和可视化途径富集,并表达表达谱作为Reactome途径的叠加。可以添加来自多个来源(包括自定义用户数据集)的蛋白质-蛋白质和化合物-蛋白质相互作用,以扩展途径。可以将路径图和分析结果显示下载为可编辑的图像,易于阅读的报告以及几种适合计算重用的标准格式的文件。通过基于代表性状态转移(REST)的内容服务和Neo4J图形数据库,可以以编程方式获得Reactome内容。IL-1至IL-38的信号传导途径在“白介素信号传导”途径中被分级分类。所使用的分类主要来自Akdis等人。
结论
向Reactome中添加一组完整的已知人类白介素,它们的受体以及与免疫功能相关方面的注释相关的既定信号通路,为有关这一重要家族的信息提供了重要的计算可访问资源。随着新发现被公认是该领域的共识,可以轻松扩展此信息。未来的主要目标是增加由白介素信号传导诱导的基因表达变化的覆盖范围。










































 京公网安备 11010802027423号
京公网安备 11010802027423号